NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
Registration now open for NCBI Discovery Workshops September 4-5 at NLM
Registration is now open for the two-day Discovery Workshops to be offered on September 4 -5, on the NIH campus in Bethesda, Maryland. The course is free and is open to anyone interested in NCBI resources. The workshops provide hands-on experience exploring practical examples using tools and databases on the NCBI website. The four workshops are Sequences, Genomes, and Maps; Proteins, Domains and Structures; NCBI BLAST Services; and Human Variation and Disease Genes. For more information see the Discovery Workshops page, which also includes a registration link.
1000 Genomes Dataset Browser
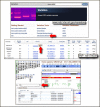
The new 1000 Genomes Browser shows variants, genotypes, and supporting sequence read alignments produced by the 1000 Genomes project. The genotype data are based on the Phase 1, March 2012 set and the variation (NCBI SNP) data are from SNP build 135. The 1000 Genomes browser is accessible from the new NCBI Variation page (Figure 1) that also provides links to other NCBI variation resources including SNP, dbVar, dbGaP, the Variation Reporter, Clinical Remap, and the Phenotype Genotype Integrator. The graphical portion of the genome browser is based on the NCBI graphical sequence viewer (GSV) and offers the familiar features and functions of the GSV. A table of genotypes organized by 1000 Genome populations is shown under the sequence viewer. A summary genotype frequency shows in the table for each population. The population sections can be expanded to show individual level genotypes.
The browser initially opens with an expanded view of human chromosome1 (Figure 1, Top panel). The search function on the right-hand side of the browser allows searches for RefSNP accession numbers, gene names, and chromosome positions. The search results show a list of matching items. Clicking on one of these items jumps the display to that position in the genome browser. The browser also allows scrolling either through the Scroll Region arrows on the table of genotypes or through the navigation controls in the graphical portion.
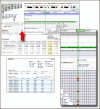
NCBI SNP records that are included in the 1000 Genomes data also link directly to the corresponding position in the browser. Figure 2 shows the region containing a SNP (rs8176058) in the Kell blood group antigen gene (KEL). For any region, the individual next-generation sequencing reads can be selected and loaded into the graphical browser through the expandable Subjects dialog box (Figure 2, Bottom panel, left). The Subjects dialog allows selecting individuals by population as well as filtering by the characteristics of the next-generation data. A portion of the aligned exome-sequencing reads for the heterozygous Toscan individual, NA20507, is shown on the left-hand side of the bottom panel of Figure 2.
The data from the 1000 genomes project are representative of the increasing importance and presence of “big data” at the NCBI. Currently these data and associated metadata are stored in many different databases at the NCBI including the Sequence Read Archive (SRA), SNP, BioSample, and BioProject. The 1000 Genomes Browser provides a simple and powerful single interface to complex and very large sets of data and metadata that comprise the 1000 Genomes project.
PubMed News
PubMed Send to Citation Manager and Favorites
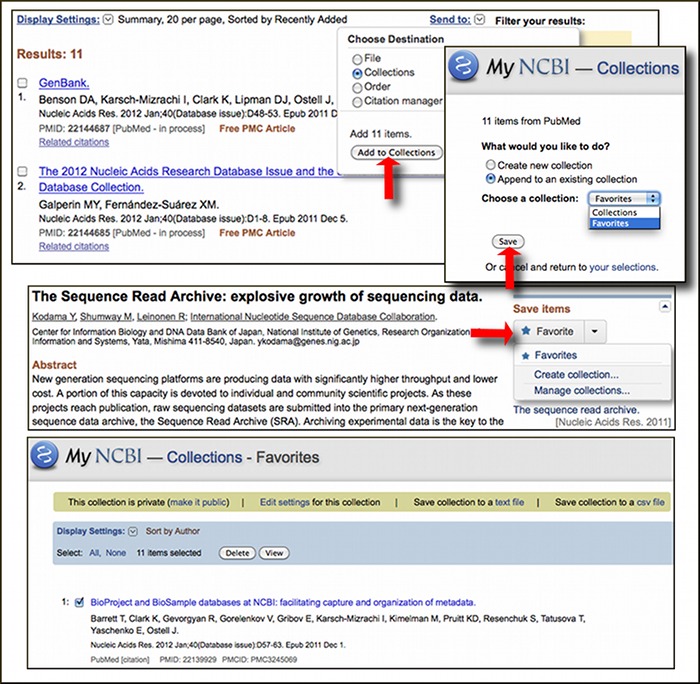
PubMed now offers the ability to download citations for use in citation manager software such as Endnote, RefWorks or other bibliography program through the "Send to" menu. The PubMed Technical Bulletin has more details on using this feature.
Abstracts in PubMed also now include a "Save items" section that will provide easy way to add items of interest to a My NCBI collection. If you are signed in to My NCBI clicking the "Favorite" button adds the citation to a new My NCBI collection, Favorites. You can add multiple items to My Collections, including Favorites, in My NCBI through the "Send to" menu in the upper right of search result displays. For more information on My NCBI and My Collections please visit My NCBI Help on the NCBI Bookshelf.
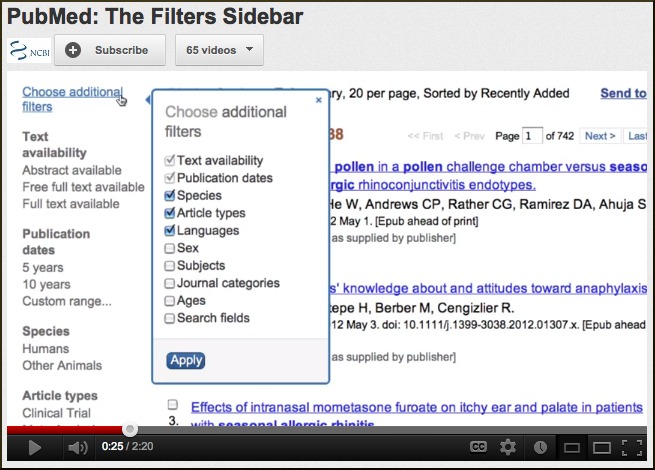
PubMed Filter Sidebar
PubMed now has a Filter Sidebar in the PubMed results. The useful features of the popular Limits page have been made more visible by placing them in this Filter Sidebar and should make it easier to refine PubMed search results. For more information, please see the NLM Technical Bulletin. A new video on NCBI’s YouTube Channel also demonstrates this useful new addition to PubMed searching.
BLAST News
New Microbial Genomes BLAST Service
A new microbial BLAST service is now live. The service is easier to use and has the familiar format and features of the standard BLAST services at NCBI including the ability to select of taxonomic categories using an auto-complete "Organism" box and to include or exclude multiple taxonomic categories. Other standard features of the BLAST pages such as "Edit and Resubmit" and the ability to optimize for a specific search are also included. For nucleotide databases the search sets have also been divided into Complete and Draft genomes.
Article on Primer-BLAST Published
An article describing Primer-BLAST, NCBI's PCR primer designing service, is now available in BMC Bioinformatics.
Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden T. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012 Jun 18;13(1):134. PubMed PMID: 22708584.
BLAST Programming Interface: End of OLD BLAST=true option
Beginning Sept. 10, 2012, the BLAST service will ignore the OLD_BLAST parameter in posted URLs. We are removing this old and little-used option to prepare for upcoming enhancements to the BLAST service later this year. Setting OLD_BLAST=true produces an older version of the BLAST HTML results that a few people have used for automated processing (parsing) of results. NCBI BLAST supports a number of different and more stable parsable formats. These include XML, tabular reports and ASN.1. For more details, please see BLAST Developer Information and links on that page.
DELTA-BLAST Service and Article
As described in the April 2012 NCBI News Domain Enhanced Lookup Time Accelerated BLAST (DELTA-BLAST) included in the BLAST 2.2.26+ release, offers a more sensitive protein-protein BLAST search by performing a position specific score matrix search using results from an initial conserved domain search. A paper in Biology Direct describes the DELTA-BLAST algorithm and discusses its enhanced sensitivity compared to other methods.
Boratyn GM, Schaffer AA, Agarwala R, Altschul SF, Lipman DJ, Madden TL. Domain enhanced lookup time accelerated BLAST. Biol Direct. 2012 Apr 17;7(1):12. PubMed PMID: 22510480.
The protein BLAST web service offers DELTA-BLAST as a Protein BLAST program selection option on the Basic Protein BLAST service.
DELTA-BLAST improves the sensitivity and selectivity of most protein searches that have strong conserved domain results. Figure 3 shows the differing alignments, scores, and expect values for the same match with standard protein blast, blastp (RID: 0E4F72U7013, Figure 3,Top panel) and DELTA-BLAST (RID: 0E5RMA6X016, Figure 3, Bottom panel). Both of these searches use the human hemoglobin subunit beta protein (NP_000509) as a query against bacterial sequences from the NCBI RefSeq protein database. Standard protein BLAST finds a globin protein (YP_485375) from the purple non-sulfur bacterium Rhodopseudomonas palustris HaA2 with an expect value of 1 X 10-4. In these results the same expect value is found for some non-globin sequences including an aspartate kinase, an amino peptidase, and a succinate-semialdehyde dehydrogenase. In addition the blastp alignment does not match the conserved histidine (position 93 in NP_000509) that is part of the heme wbinding site in the human hemoglobin domain and structure (Figure 3, Middle panel). The blastp alignment also inserts a gap in the conserved alpha helix in this region. In contrast, DELTA-BLAST finds the same protein but with a much better expect value of 3 X 10-27, thus easily segregating the hit from the non-globin proteins found in the blastp search. Moreover, the alignment now corresponds to the globin conserved domain, matching the conserved histidine and preserving the secondary structure block.
DELTA-BLAST is an important new addition that extends the capabilities of the NCBI BLAST service and produces more accurate alignments and more discriminating statistics by using conserved domain information in the initial search.
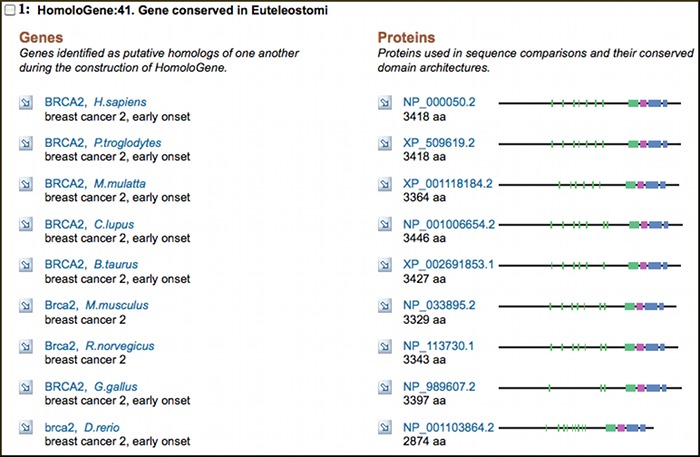
New HomoloGene Build: Rhesus macaque now included
HomoloGene, the NCBI resource that identifies and clusters homologous genes, transcripts and proteins for selected eukaryotes, has a new build (Build 66). With this build, HomoloGene for the first time includes genes and sequences for the Rhesus monkey (Macaca mulatta). The new build also includes updated annotations for human, chimpanzee, dog, cow, mouse, rat, chicken, zebrafish, fruitfly, yeast, arabidopsis, and rice. HomoloGene data are available from the NCBI FTP site.
Microbial Genomes Update
Ninety-two finished microbial (archaeal and bacterial) complete genome sequences were released for 90 microbial strains (7 archaea and 83 bacteria) from April 2012 through June 2012. These include three complete plasmid sequences and 89 chromosome sequences. The original sequence data files submitted to the International Sequence Database Collaboration (INSDC) are available in the Bacteria directory in the genomes area of the GenBank FTP site. RefSeq versions were released for a selected set of 391 of the complete INSDC microbial genome sequences for 387 microbial strains during the same period. These are available from the /genomes/Bacteria directory on the FTP site.
In addition, data from 754 microbial whole genome-shotgun (WGS) sequencing projects were added to the INSDC during this period. The original submitted files are available in the Bacteria_DRAFT directory in the GenBank genomes area. RefSeq provisional versions of 84 WGS microbial projects were released in the /genomes/Bacteria_DRAFT area of the FTP site.
All GenBank and RefSeq microbial genomes are incorporated in the NCBI integrated Entrez search and retrieval system and the BLAST sequence similarity search service.
GenBank News
GenBank release 190 is available through the NCBI web and FTP sites. The current release incorporates data available as of June 15, 2012 and, with the whole-genome shotgun portion, contains 428,920,607,871 bases from 236,206,989 sequence records. Release notes describe the current state of data and upcoming changes. The GenBank page provides more information on the database content and scope as well as submission information.
RefSeq News
RefSeq Release 54 is available through Entrez, BLAST, and from the RefSeq FTP area. The current release includes 21.9 million Reference Sequence records from 17,605 different species or strains. The RefSeq release notes provide more detailed information.
GRC Plans New Human Genome Build and Requests Input
The Genome Reference Consortium (GRC), which produces assemblies that are the basis for NCBI Reference assemblies for human, mouse, and zebrafish, is planning a new build of the human genome (GRCh38) for summer of 2013. Anyone who has questions, concerns, or input, may submit these on the GRC contact form. The GRC blog provides insights into the complexities and the process of updating, correcting, and representing the human genome.
NCBI Now Offers IPv6 Access
The NCBI website now supports the new six-byte Internet Protocol addresses (IPv6) for HTTP access as well as data downloads using FTP, Aspera, and RSync. The World IPv6 Launch site has additional information on the transition to IPv6.
Keeping Up with NCBI
Seventeen topic-specific mailing lists are available that provide email announcements about changes and updates to NCBI resources including dbGaP, BLAST, GenBank, and Sequin. The various lists are described on the Announcement List summary page. Subscribe to the NCBI Announce list to receive updates on the NCBI News.
Twenty-six RSS feeds are now available from NCBI including news on PubMed, PubMed Central, NCBI Bookshelf, LinkOut, HomoloGene, UniGene, and NCBI Announce.
NCBI’s Facebook page and Twitter feed also provide updates on NCBI resources.
Send comments and questions about NCBI resources to info@ncbi.nlm.nih.gov, or call 301-496-2475 between the hours of 8:30 a.m. and 5:30 p.m. EST, Monday through Friday.
- Registration now open for NCBI Discovery Workshops September 4-5 at NLM
- 1000 Genomes Dataset Browser
- PubMed News
- BLAST News
- New HomoloGene Build: Rhesus macaque now included
- Microbial Genomes Update
- GenBank News
- RefSeq News
- GRC Plans New Human Genome Build and Requests Input
- NCBI Now Offers IPv6 Access
- Keeping Up with NCBI
- NCBI News, July 2012 - NCBI NewsNCBI News, July 2012 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...