NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 1997.
Nature of the RNA Template
Retroviruses differ from other viruses in that each virion contains two complete copies of the single-stranded RNA genome. It has been proposed that retroviruses copackage two genomic RNAs to increase the probability of successful DNA synthesis: If one of the two RNAs is broken, RT can switch templates and copy the copackaged RNA, thereby permitting DNA synthesis through the site of the lesion (Coffin 1979). Only a single DNA is believed to be formed in an infectious retroviral particle. Hence, retroviruses are not truly diploid in the usual sense: Any given position in the progeny DNA contains genetic information from only one of the two parental RNAs (Panganiban and Fiore 1988; Hu and Temin 1990a; Jones et al. 1994). Nevertheless, the pseudodiploid nature of the retroviral genome has important implications for the generation of diversity in retroviruses, as discussed below.
Retroviral genomic RNA is a product of the host RNA synthesis machinery, and as such, the viral RNA genome has the structural features of a cellular messenger RNA, including a 7-me-G cap and a polyadenylated 3′end. Within retroviral particles, the two plus-strand genomic RNAs are noncovalently joined at multiple points, most strongly near their 5′ends, in what is called a dimer linkage structure (Bender et al. 1978; Murti et al. 1981; Darlix et al. 1990; Awang and Sen 1993; Muriaux et al. 1995). A host-derived tRNA is annealed via a segment at its 3′end to a complementary region near the 5′end of the viral genome (Taylor 1977). Virions also contain a variety of nonprimer host tRNAs (∼50–100 copies per virion) and variable amounts of other host RNAs, some of which can be reverse-transcribed (Dunn et al. 1992); however, none of these ancillary host RNAs are known to serve any important function in retroviral replication (Linial and Miller 1990).
Reverse transcription takes place in a ribonucleoprotein complex that includes not only RT and the diploid RNA genome, but other viral proteins as well. It would appear that this complex provides a particularly favorable environment for reverse transcription; it is likely that NC specifically facilitates the synthesis of viral DNA. NC can be thought of as the retroviral equivalent of prokaryotic single-stranded binding protein (SSB), and, like SSB, NC has been reported to both bind single-stranded nucleic acids and promote nucleic acid annealing (Schulein et al. 1978; Smith and Bailey 1979; Karpel et al. 1987; Prats et al. 1988, 1991; Darlix et al. 1990; Weiss et al. 1992; see Chapter 2. NC also has a role in retroviral RNA dimerization and packaging (Meric and Spahr 1986; Gorelick et al. 1988, 1990, 1993, 1996; see also Chapter 3. The substrate for reverse transcription is a protein RNA-complex; the presence of NC may enhance the processivity of reverse transcription and template switching (Fütterer and Hohn 1987; Allain et al. 1994; Peliska et al. 1994). The role in reverse transcription (if any) of the other virion proteins that appear to be part of the ribonucleoprotein complex is less clear. There is some evidence that viral protease (PR), which functions in the maturation of newly assembled virions, may be required during preintegrative phases of retroviral replication: It is possible that its role is to perform additional cleavages of mature NC to produce a form of NC that specifically promotes reverse transcription (Nagy et al. 1994).
The ends of retroviral genomic RNA contain sequence elements important for reverse transcription (Fig. 1). These include the direct repeats, termed R for repeated sequences, that lie at the 5′and 3′ends of the RNA immediately adjacent to the 5′cap and 3′ poly(A) tail (Coffin and Haseltine 1977; Haseltine et al. 1977; Schwartz et al. 1977; Stoll et al. 1977; Coffin et al. 1978). Sequences that are unique to each end of the viral RNA and are adjacent and internal to the R sequences have been called U5 (unique to the 5′end) and U3 (unique to the 3′end). Additional sequences, the PBS and the PPT, are required either to position or to become the RNA primers for minus- and plus-strand DNA synthesis. The overall arrangement of these sequence elements is (from the 5′cap): cap, R, U5, PBS... PPT, U3, R, poly(A). The ends of the unintegrated viral DNA genome are defined by the sites where RNase H removes the specific plus-strand and minus-strand primers. These cleavage sites also define the interior boundaries of U3 and U5 in the product DNA. For most retroviruses, cleavages occur at the junction between the RNA primer and the first DNA base synthesized by RT; however, in the case of human immunodeficiency virus type 1 (HIV-1), RNase H cleavage leaves the 3′-most RNA base of the tRNA that was used as the minus-strand primer on the end of the DNA (Whitcomb et al. 1990; Pullen et al. 1992; Smith and Roth 1992). Sequences that are termed “Psi” (ψ) or alternatively “E” are required for the specific encapsidation of viral RNAs and are thus required (albeit indirectly) for reverse transcription, because they are responsible for delivering the RNAs to the virion particle (Rein 1994). All of these sequence elements are maintained in and, in the case of U3 and U5, duplicated in viral DNA. The duplication of U5 and U3 sequences yields the characteristic LTR (5′-U3, R, U5-3′) found at both ends of retroviral DNA. Generating the linear 2-LTR form of retroviral DNA is necessary both to establish DNA ends that are suitable substrates for integration (see Chapter 5 and to regenerate regulatory sequences (the promoter and polyadenylation sites) that are lost when the provirus is transcribed into genomic RNA.
The cis-acting reverse transcription sequences vary among retroviruses. For example, the R region can be as small as 12 bases or as large as 229 bases. Although longer repeated sequences enhance strand transfer in vitro (Luo and Taylor 1990; Arts et al. 1994), the additional sequences that make the R regions of some viruses longer than others are probably not required for reverse transcription per se but are retained because the sequences have additional purposes: The TAR region of HIV-1 is an important example (Muesing et al. 1987; Berkhout et al. 1995; see Chapter 6. The PBS and PPT sequences vary less in length than in composition, reflecting the specific tRNA primer used and possibly differences in the interactions with different retroviral RTs.
RT in the Virion
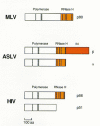
Retroviral particles are formed as assemblies of polyprotein precursors (see Chapters 2 and 7). In most retroviruses, RT is expressed as part of a Gag-Pol polyprotein that contains, in addition to Gag sequences, the viral enzymes PR and integrase (IN) (Jamjoon et al. 1977; Oppermann et al. 1977; Kopchick et al. 1978; for review, see Eisenman and Vogt 1978; Levin et al. 1993). The ratio of Gag to Gag-Pol is controlled at the translational level either by reading through the Gag termination codon (nonsense suppression) or by frameshifting. A special tRNA is not required for either type of suppression; readthrough depends on interactions between elements in the viral RNA and the normal translational machinery of the host cell. Gag self-assembles; the Gag-Pol polyprotein is incorporated into virions because of Gag-Gag interactions (see Chapters 2 and 7). The organization of the Gag-Pol polyprotein places the Pol portion of the Gag-Pol polyprotein in the interior of the virion. Mature RT is liberated from these precursors by the viral PR; for most retroviruses, this occurs during, or shortly after, viral budding. The sites of the proteolytic cleavages that generate mature RT vary from retrovirus to retrovirus, resulting in RTs with different subunit compositions (Fig. 3). Each mature virion contains approximately 100 molecules of RT.
It was recognized only quite recently that in foamy viruses, RT is not synthesized as a Gag-Pol polyprotein, but it is synthesized separately from Gag. In foamy viruses, RT is translated from a spliced RNA (Yu et al. 1996; Enssle et al. 1996). Hence, whereas the RTs of most retroviruses assemble into virions as segments of Gag-Pol polyprotein precursors, the inclusion of RT into foamy virions must involve a distinct, as of yet uncharacterized, mechanism.
RT can be readily liberated from isolated virions by gentle detergent permeabilization of virion envelopes. This suggests that RT is not firmly attached to the viral genome in the virion core; however, the association of RT with an infecting virion is essential. Reverse transcription of the viral genome will not occur when RT-deficient virions enter cells even if RT is provided in trans (Ansari-Lari and Gibbs 1996).
Complete viral DNA can be synthesized—albeit inefficiently—in purified virions in the so-called “endogenous reaction,” in which purified virions are permeabilized with a mild detergent and incubated in a reaction mixture containing deoxyribonucleotides. The endogenous RT reaction has had an important role in the history of retrovirology: It was this assay that was first used to demonstrate the existence of RT (Baltimore 1970; Temin and Mizutani 1970). In addition, it has been used to help decipher the complexities of copying the viral genome into DNA. Although the reaction is relatively inefficient, it is possible to make small amounts of full-length viral DNA in vitro, which demonstrates that all of the components (except the dNTPs) required for the reverse transcription reaction are contained in the virion (Haseltine et al. 1976; Rothenberg and Baltimore 1976; Rothenberg et al. 1977; Gilboa et al. 1979b; Mitra et al. 1979; Boone and Skalka 1981a). Since adding nucleotides to detergent-permeabilized mature virions initiates reverse transcription in the endogenous reaction, the absence of nucleotides is presumably the primary reason reverse transcription is not initiated in extracellular virions.
The conditions required for producing complete reverse transcripts in the endogenous reaction are exquisitely sensitive to the concentration of detergent (usually Nonidet P-40, NP-40) that is present in such in vitro reactions (Rothenberg and Baltimore 1976; Rothenberg et al. 1977; Boone and Skalka 1981a). With too little detergent, overall DNA synthesis is poor, whereas with too much detergent, some early intermediates are formed, but strand transfer is inefficient and synthesis of the plus strand of DNA does not occur. This sensitivity to detergent suggests that an intact virion core is required for efficient elongation and strand transfer. Whether the core serves only to confine the components necessary for reverse transcription at high concentrations or whether the structure of the core specifically promotes proper synthesis in more complex ways is unclear. Furthermore, how well the structures present in detergent-disrupted virion reactions mimic the intracellular forms that carry out the reactions in vivo is also uncertain. There are significant differences in the efficiency of reverse transcription in vitro and in vivo; even under the best conditions in vitro, only a few percent of the product DNA is full-length double-stranded DNA (Gilboa et al. 1979b; Boone and Skalka 1980), and most of the DNA seen in vivo is often close to full length.
The Nature of the Intracellular Reverse Transcription Complex
Although there is evidence of limited DNA synthesis in HIV-1 virions prior to the entry of the virion into the target cell (Lori et al. 1992; Trono 1992; Zack et al. 1992; Zhu and Cunningham 1993), reverse transcription in general appears to be activated by entry of the viral core into the cytoplasm of the target cell. The structure and/or organization of the Gag-Pol polyprotein (or its relationship to viral RNA) presumably prevents reverse transcription from occurring before the virion is budded from the cell. There is no absolute requirement for reverse transcription to occur only after a virion is released. In hepadnaviruses, extensive reverse transcription occurs before virions are released from producer cells; this led to their initial classification as DNA viruses.
The process of viral entry and uncoating remains one of the most mysterious steps in the life cycle of the virus (see Chapter 3. A major difficulty is that the majority (usually >99%) of the virions applied to a cell do not initiate productive reverse transcription—either because they are damaged or because the process of entry is inefficient—and so monitoring the status of the virion proteins and genomes early after infection is risky, potentially revealing more about the inactive virions than about the active virions. Genetic experiments suggest that Gag proteins are important in proper activation of virion-associated reverse transcription. Mutants altered in matrix (MA), murine leukemia virus (MLV) p12, and the capsid (CA) often produce normal levels of virions but are unable to initiate reverse transcription in an acutely infected cell (Schwartzberg et al. 1984; Crawford and Goff 1984; Hsu et al. 1985; Strambio-deCastillia and Hunter 1992; Reicin et al. 1996). These particles mediate normal reverse transcription in vitro when exposed to detergents and triphosphates, which shows that the viral RNA template and RT are not defective. Further evidence of the involvement of Gag in these early events is provided by studies of HIV-1 virions. A host protein, cyclophilin A, which binds to HIV-1 CA (Luban et al. 1993), is present and is required for proper entry or uncoating (Franke et al. 1994; Thali et al. 1994). The cyclosporin drugs, which bind to cyclophilins and disrupt the Gag-cyclophilin interaction, block some early event before viral DNA synthesis begins (Braaten et al. 1996). In the case of HIV-1, other viral proteins have been implicated, at least indirectly, in reverse transcription. It has been reported that Nef-deficient viruses produce reduced amounts of viral DNA (Schwartz et al. 1995) and that Vif has a role in reverse transcription (Sova and Volsky 1993; von Schwedler et al. 1993; Goncalves et al. 1996).
In actively growing cells, reverse transcription can ordinarily proceed in an uninterrupted fashion through the steps outlined above. DNA synthesis can be essentially completed in the cytoplasm; viral nucleoprotein complexes isolated from the cytoplasm of infected cells are competent to carry out the integration of the viral DNA into target DNA in vitro (Brown et al. 1987; Bowerman et al. 1989; see Chapter 5. If the host cell is serum starved before infection, reverse transcription is initiated but not completed, perhaps because the pool of dNTPs present in the cytoplasm is insufficient (Chen and Temin 1982; O'Brien et al. 1994). However, the complex is quite stable, and reverse transcription can be completed at later times if the cells are refed with serum (Fritsch and Temin 1977; Varmus et al. 1977; Zach et al. 1990, 1992).
Avian leukemia virus (ALV) may be an exception to the general rule that reverse transcription is completed in the cytoplasm. Much of the ALV DNA in the cytoplasm appears to be incomplete (Varmus et al. 1978; Lee and Coffin 1991). For ALV, the completion of DNA synthesis—both minus and plus strands—may require transit into the nucleus. This delayed completion might confer advantages to the virus. It might limit the generation of the aberrant DNA products produced by autointegration (Chapter 5, and it might also help to ensure an adequate supply of dNTPs. For other viruses, complete double-stranded viral DNA can be isolated from infected cells starting about 4 hours postinfection, although the period of time required may vary widely depending on the choice of host cell and virus (Varmus and Swanstrom 1982, 1985; Kim et al. 1989).
The nucleoprotein complex within which reverse transcription occurs appears to resemble, at least superficially, the core of an extracellular virion (Bowerman et al. 1989). In particular, some of the Gag proteins and the Pol proteins, RT and IN, remain associated with the RNA and DNA for some time in the infected cells, although there is some controversy as to which proteins are retained, and it is possible that different retroviruses differ in this regard. At present, MLV complexes have been reported to contain, at a minimum, CA (Bowerman et al. 1989); HIV-1 complexes seem to contain MA but not CA (Farnet and Haseltine 1991; Bukrinsky et al. 1993) and probably Vpr as well. The evidence for the presence of some of these components is genetic as well as biochemical.
Host factors may have a role in reverse transcription in vivo. Fv1 restriction is associated with the presence of specific sequences in the CA protein of MLV. In a restrictive host cell, the efficiency of viral infection is reduced, and the defect is associated with a deficiency in integration which may or may not be the result of defects in the late stages of reverse transcription. The identification of the Fv1 locus as a defective endogenous retrovirus has not provided a clear solution to this question (Best et al. 1996; see Chapter 5. The interaction of the CA of HIV-1 with cyclophilin (and a possible effect on reverse transcription) has already been discussed; however, this interaction appears to occur only in HIV-1. It is also possible that viral components may be joined by, or interact with, other host factors from either the virion-producing cell or the infected cell. Thorough characterization of the intracellular viral nucleoprotein complex has been confounded not only by the relatively low abundance of these complexes as compared to host-cell nucleoprotein assemblies, but also by the fact that for retroviruses, the ratio of particle to infectivity is low and infected cells probably contain a relatively large proportion of products derived from defective virions. It is not known if, when, or to what extent virion proteins such as NC and RT are lost from the core complex. Whether replication is aborted because of the loss of essential components of the nucleoprotein complex or for other reasons is unknown, in part because it is not presently possible to separate replication-competent viral nucleoprotein complexes from defective ones.
Process of Reverse Transcription
Much of what is known about the process of reverse transcription was learned by identifying replication intermediates synthesized in either endogenous or reconstituted reactions and by analyses of viral DNA isolated from actively infected cells (Haseltine et al. 1976, 1979; Weinberg 1977; Shank et al. 1978; Varmus et al. 1978; Boone and Skalka 1980, 1981a,b; Gilboa et al. 1979b; Borroto-Esoda and Boone 1991). The basic features of this model for reverse transcription have remained essentially unchanged since they were first proposed; experimental support for the model has been extensively reviewed (Fig. 2) (Gilboa et al. 1979a; Varmus and Swanstrom 1982, 1985; Coffin 1990; Whitcomb and Hughes 1992; Champoux 1993; Telesnitsky and Goff 1993a).
Priming/tRNA Choice
RT, like many other DNA polymerases, cannot initiate DNA synthesis without a primer strand to extend and a template strand to copy. The primer used by RT to initiate minus-strand DNA synthesis is a host-encoded tRNA. The host tRNA is partially unfolded from its native structure so that 18 nucleotides at the 3′terminus of the tRNA can be base-paired to a specific complementary sequence, termed the PBS, on the genomic RNA. Different tRNA primers are used by retroviruses of different genera; for example, tryptophan for the avian sarcoma/leukosis viruses (ASLVs) (Harada et al. 1975), proline for the mammalian C-type viruses (Peters et al. 1977; Harada et al. 1979) (and in some cases glutamine for endogenous mammalian viruses), histidine for a fish retrovirus (Holzschu et al. 1995), and lysine(3) for both mouse mammary tumor virus (MMTV) (Peters and Glover 1980) and HIV-1 (Ratner et al. 1985; Jiang et al. 1993). Retroviral virions contain a large excess of free tRNA, probably about 50–100 molecules per virion. The relative abundance of different tRNAs present in the virion can be significantly different from the distribution of tRNAs present in the host cell in which the virion was assembled. In such cases, it is likely that the RT segment of the Gag-Pol polyprotein participates in recruiting specific tRNAs for packaging (Peters and Hu 1980; Mak et al. 1994). All mature RTs show some affinity for binding tRNAs, although the selectivity shown by the RT for specific tRNAs differs for different retroviruses. Some mature RTs appear to bind preferentially to their cognate primer tRNAs (e.g., HIV-1 and ALV) (Panet et al. 1975; Haseltine et al. 1977; Hu and Dahlberg 1983; Garret et al. 1984; Barat et al. 1989, 1991; Sarih-Cottin et al. 1992). There are, however, mature RTs (e.g., MLV) that exhibit only modest selectivity for any particular tRNA (Panet and Berliner 1978). The ultimate determinant of which tRNA is used for priming is the PBS sequence itself, with which the tRNA must pair. This statement is to some extent self-fulfilling. One of the strands of the PBS sequence in the viral DNA is derived from copying a portion of the tRNA; in this way, the sequence of the tRNA primer can, after replication, become a part of the sequence of the PBS.
Many retroviruses are relatively flexible in their choice of primers. In the case of murine retroviruses, a naturally occurring viral variant and several targeted PBS mutants have shown that alternative tRNA primers can be used (Colicelli and Goff 1986; Lund et al. 1993), and various tRNAs can serve as primers in reconstituted reactions in vitro (Kohlstaedt and Steitz 1992). However, for both HIV-1 and ALV, the natural primer appears to confer some replicative advantage. HIV mutants with PBS sequences complementary to tRNA (phenylalanine) or tRNA (lysine 1,2) could be passaged in tissue culture. However, wild-type PBS sequences were restored after several rounds of replication (Li et al. 1994; Wakefield et al. 1994; Das et al. 1995). ALV mutants with PBS sequences complementary to a number of different tRNAs were also able to replicate. Although all of these mutants eventually reverted to the wild-type tryptophan PBS, the rate of reversion differed for different mutants (Whitcomb et al. 1995).
The tRNA primer is properly annealed to the PBS in the absence of a functional PR, which shows that cleavage of the Gag and/or Gag-Pol polyproteins is not essential for the annealing reaction. The role(s) of individual components of the polyproteins is less well defined; however, it has been suggested that both NC and RT play a part in the process (Prats et al. 1988; Khan and Giedroc 1992; Barat et al. 1993). A complicating factor is that the rules may be different for different retroviruses. The mature RT of the ASLVs binds its cognate primer selectively, but the RT of MLV is much less selective (Panet et al. 1975; Haseltine et al. 1977; Panet and Berliner 1978; Cordell et al. 1979). It therefore may not be surprising that in an MLV mutant that lacks the Pol proteins, the tRNA primer is still properly bound to the PBS; the corresponding ASLV mutants do not put the tRNA primer on the PBS (A. Rein, pers. comm.). It has been suggested that sequences outside of the PBS in genomic RNA and host tRNA interact in important ways (Aiyar et al. 1992, 1994; Kohlstaedt and Steitz 1992; Leis et al. 1993; Arts et al. 1994). Recent experiments involving MLV, ASLV, and HIV-1 mutants that seem to be able to use alternative tRNAs with reasonable facility bring some of the specific ideas into question; however, there is a report that secondary mutations in the genome of HIV-1, which would be expected to enhance the interaction with a nonstandard tRNA primer, enhanced the replication of the mutant virus (Wakefield and Morrow 1996). The reversion of ALV and HIV mutants with altered PBS may also reflect not only a preference for tRNAs present at relatively high concentrations in virions (Whitcomb et al. 1995), but also an attempt, by the virus, to optimize secondary interactions between the viral genome and the tRNA primer.
Minus-strand Synthesis and the First Strand Transfer
The synthesis of minus-strand DNA is initiated from the tRNA primer which is based-paired at the PBS near the 5′end of the genomic RNA. Copying of the genomic RNA template up to the 5′cap results in the formation of a short minus-strand DNA usually called –sssDNA. Continued minus-strand DNA synthesis requires a strand-transfer reaction that allows the 3′end of the genomic RNA to serve as a template (Varmus et al. 1978; Gilboa et al. 1979b; Mitra et al. 1979; Swanstrom et al. 1981). The appearance of a discrete –sssDNA early in the endogenous reaction in vitro suggests that minus-strand synthesis proceeds until the 5′end of the genomic RNA is reached and that the first strand transfer occurs only after the complete –sssDNA product has been formed. However, although –sssDNA is found in cells infected with certain RNase H mutant viruses, discrete-length –sssDNA is not detectable in cells infected with wild-type retroviruses, indicating that the first strand transfer occurs rapidly in vivo (Lee and Coffin 1991; Blain and Goff 1995). It is conceivable that, in vivo, the first strand transfer could occur after only a portion of R has been copied. Strand transfer can occur from internal template regions in reconstituted reactions (DeStefano et al. 1992b, 1994a). In genetic studies involving viruses or vectors whose 5′ and 3′R sequences were not identical, 3′R sequences were sometimes observed in the DNA product, suggesting that first strand transfer can occur before all of the 5′R sequence is copied (Lobel and Goff 1985; Ramsey and Panganiban 1993; Klaver and Berkhout 1994). Whether these early jumps represent a form of copy choice recombination or whether they are simply a portion of the normal first strand transfer reactions is not clear. However, at least in the case of Moloney MLV (Mo-MLV), the vast majority of first strand transfers appear to occur after the completion of –sssDNA (Kulpa et al. 1997).
The RNase H activity of RT is required for the first strand transfer. Viruses harboring mutations in RT that inactivate RNase H are noninfectious (Repaske et al. 1989; Tanese et al. 1991; Tisdale et al. 1991). In endogenous reactions with purified virions that contain RNase-H-defective RTs, –sssDNA remains in the form of an RNA-DNA hybrid and the extent of first strand transfer is substantially diminished (Tanese et al. 1991; Telesnitsky et al. 1992). All studies that have used purified model templates to study strand transfer in vitro have reported defects in template switching when RNase-H-deficient RTs were used in the reaction (Luo and Taylor 1990; Buiser et al. 1991; Garces and Wittek 1991; Peliska and Benkovic 1992; Ghosh et al. 1995).
Even when the RT retains its full RNase H activity, the 3′end of –sssDNA is likely to exist, at least initially, as an RNA-DNA hybrid. When RT reaches the 5′ end of the RNA genome, the geometry of the enzyme– primer template complex dictates that the RNase-H-active site lies 15–18 bases “behind” the 5′end of the template. In vitro RT preferentially cleaves an RNA template at approximately this distance from the 3′end of a DNA primer (Wöhrl and Moelling 1990; Furfine and Reardon 1991; Fu and Taylor 1992; Gopolakreshnan et al. 1992). Hence, it may be difficult for RNase H to cleave the 5′portion of the viral genome, and some mechanism in addition to RNase H digestion may be required to prepare DNAs for template switching. It has been suggested that the first jump could involve a transient melting of the residual RNA:DNA segment to unmask the 3′end of –sssDNA (Collett et al. 1978). Alternatively, terminal RNA remnants might be displaced following an initial pairing of minus-strand DNA sequences internal to the terminal RNA:DNA duplex to complementary acceptor template sequences followed by branch migration to displace genomic RNA remnants (Champoux 1993; DeStefano et al. 1994a). Although HIV-1 RT initially cleaves an RNA template 15–18 bases from the 3′end of a DNA primer strand, the enzyme is capable, with longer incubation, of making additional cuts approximately 8 bases from the end of the primer (Oyama et al. 1989; DeStefano et al. 1991a; Peliska and Benkovic 1992; Ghosh et al. 1995). Such fragments may be small enough to permit efficient strand transfer, particularly in the presence of NC.
Once the first jump has occurred, the 3′end of the minus strand can be extended and long minus-strand DNA products are synthesized. If endogenous reactions are carried out in the presence of actinomycin D, DNA-dependent DNA synthesis does not occur, and only minus-strand species are formed. The size of the longest of these species suggests termination occurs near the PBS region of the genomic RNA. This finding has been explained either as an inability of RT to displace nucleic acids annealed 5′to the PBS or, more plausibly, to the absence of any intact RNA remaining 5′to the PBS, since if –sssDNA was made from both RNA genomes, the 5′ends of both RNA strands would have been degraded by RNase H.
Plus-strand Initiation
The site where plus-strand synthesis is initiated and where the plus-strand primer is removed by RNase H defines both the interior border of U3 and the upstream end of the LTR (and hence, the end of the viral DNA). This means that the accurate generation and removal of the plus-strand primer is required to produce integration-competent DNA. RT produces the short RNA primer used to initiate plus-strand synthesis by making specific RNase H cleavages of the genomic RNA when it is in the form of an RNA-DNA hybrid (this hybrid is generated by minus-strand DNA synthesis). The primer derives from a region known as the polypurine tract (PPT), whose name reflects its base composition. Although the base composition is conserved, PPT sequences vary from virus to virus. In reconstituted reactions, in vitro, a given RT is able to cleave its own PPT accurately (Finston and Champoux 1984; Smith et al. 1984b; Rattray and Champoux 1989; Luo et al. 1990). RTs from some retroviruses are also able to cleave the PPTs of other viruses appropriately; however, the edges of the primer and site of initiation may not be precise or at the correct sites (Champoux et al. 1984; Pullen et al. 1993).
Although all retroviruses create a specific plus-strand primer from the PPT, some viruses—notably HIV and the ALVs—also use additional internal plus-strand primers which also derive from genomic RNA (Harris et al. 1981; Kung et al. 1981; Hsu and Taylor 1982; Charneau and Clavel 1991; Tobaly-Tapiero et al. 1991; Charneau et al. 1992). Thus, the plus strand in the product DNA can be discontinuous (Varmus et al. 1978; Kupiec et al. 1988; Miller et al. 1995). Plus-strand synthesis is believed to be continuous for MMTV and MLVs, but alternative upstream plus-strand starts are observed as minor species in Mo-MLV endogenous reactions, with DNA synthesis initiating from sequences that resemble the authentic PPT (Kung et al. 1981; DesGrolliers et al. 1982; Rattray and Champoux 1987). The role of internal plus-strand starts in replication is unclear, but they could be used to accelerate the overall rate of formation of completed DNA and/or to facilitate some kinds of genetic recombination (Katz and Skalka 1990).
The central PPT in HIV-1 appears to be critical for viral replication (Charneau et al. 1992). Unintegrated HIV-1 DNA has a central plus-strand overlap that appears to result from limited displacement of the plus-strand DNA initiated at the central PPT by plus-strand DNA that is initiated at the U3-proximal PPT. Strand displacement synthesis halts at a termination site specific for strand-displacement synthesis, and mutagenesis of this terminator impairs viral replication (Charneau et al. 1992). Mutations of the central HIV-1 PPT also block steps in replication after reverse transcription, raising the possibility that these discontinuities may be important for nuclear entry or establishment of the provirus in the host genome (Charneau et al. 1994). HIV-1 preintegration complexes that retain plus-strand discontinuities are competent to integrate in vitro, suggesting that DNA synthesis may be completed by host enzymes after integration has occurred (Miller et al. 1995).
Mutational studies of U3-proximal PPT regions have indicated that certain sequences within the PPT are particularly important in specifying RNase H cleavage. In the case of MLV, mutations at nucleotide –7 relative to the site of RNase H cleavage appear to influence the precision of cleavage in reconstituted reactions more dramatically than does any other individual mutation between –6 and +5 (Rattray and Champoux 1989). In the case of HIV-1 RT, sequences at –2 and –4 appear to be critical for precise cleavage (Pullen et al. 1993). There is, however, a real question of what defines the specificity of cleavage. Although it is a relatively simple matter to describe the effect of altering the sequence of the RNase H substrate, it is quite likely that it is the structure of the RNA:DNA duplex, or more properly of the complex between the RNA/DNA and RT, that defines the specificity of the cleavage (Jacobo-Molina et al. 1993; Boyer et al. 1994c; Arnold et al. 1995; Wöhrl et al. 1995b). On the basis of the RT structural data discussed below, these critical PPT regions are likely to contact the DNA polymerase domain of RT. However, the ability of RT to generate correct PPT cleavages on preformed RNA:DNA duplexes that contain the PPT suggests that specificity of RNase H cleavage does not result from RT pausing during DNA synthesis and that specific cleavages in the PPT can occur when RT rebinds the RNA-DNA hybrid after the minus-strand DNA complement of the region has been synthesized (Huber and Richardson 1990; Luo et al. 1990).
RT must make at least two specific cleavages in the PPT during retroviral DNA synthesis: one to generate the primer and a second when the RNA primer is removed. The initial plus-strand DNA product that results from DNA synthesis initiated at the PPT primer is termed +sssDNA. This DNA product is initiated from the 3′end of the PPT oligoribonucleotide, generated by RNase H. Copying the 5′end of the minus-strand DNA generates +sssDNA, which is terminated after a portion of the primer tRNA has been copied. The +sssDNA has a very discrete length both in vitro (Mitra et al. 1979) and in vivo (Shank et al. 1978; Varmus et al. 1978; Roth et al. 1989; Lee and Coffin 1991), suggesting that its synthesis is terminated specifically, probably at the site of the first modified base in the tRNA primer, a 1-methyl-adenosine residue that cannot serve as a template for reverse transcription (Youvan and Hearst 1979; Swanstrom et al. 1981). Using part of the tRNA primer as a template yields a short RNA-DNA hybrid that permits the removal of the primer tRNA by RNase H (Cordell et al. 1979; Omer and Faras 1982; Champoux et al. 1984; Champoux 1993).
Although, as far as is now known, RNase H always removes the entire PPT primer from +sssDNA, in at least one other situation, RNase H leaves a single RNA base attached to a cleavage product. This case is when the HIV-1 RT removes the tRNA primer from the minus-strand DNA (Kulkosky et al. 1990; Smith et al. 1990; Whitcomb et al. 1990; Pullen et al. 1992; Smith and Roth 1992). It is still unclear what defines the specificity of this cleavage; however, in HIV-2, the entire tRNA is removed (Whitcomb and Hughes 1992).
Second Jump
Copying a portion of the primer tRNA produces a DNA copy of the PBS or, more accurately, a copy of the end of the tRNA used to initiate minus-strand DNA synthesis at the 3′end of +sssDNA. According to prevailing models for reverse transcription, minus-strand synthesis proceeds through the PBS. Presumably, the tRNA primer either is displaced by minus-strand synthesis or has already been displaced by plus-strand synthesis. The two complementary PBS sequences on the plus and minus DNA strands can then serve as the complementary regions required for the second strand transfer.
This model predicts that in newly synthesized viral DNA, the plus strand of the PBS sequences is copied from the tRNA primer, and the minus strand of the PBS sequences is copied from the viral genome. If the sequence of the tRNA primer does not match the sequence of the PBS in the genome, information from the tRNA primer can be introduced into the PBS of the viral DNA via reverse transcription. This is presumably what happens when HIV-1 and ASLV mutants in which the PBS has been changed to pair with alternate tRNA primers revert to wild type (Li et al. 1994; Wakefield et al. 1994; Das et al. 1995; Whitcomb et al. 1995). A specific example that suggests the existence of heteroduplexes with different sequences on the plus and minus strands of the PBS was described by Berwin and Barklis (1993). In these experiments, a single base change in the PBS enables an MLV variant to grow in a specific cell type. This alteration results in a mismatch between the PBS and the tRNA primer. From a single infected cell, progeny can be derived that contain proviruses whose PBS sequences match both the sequences originally present in the viral genome and the tRNA primer. This suggests that the viral DNA that was integrated contained a mismatch at the PBS and that the replication of the host genome occurred before this mismatch could be corrected.
Apparently, the second DNA strand transfer does not generally occur until after +sssDNA has been completed and the minus strand has extended through the PBS, at least for spleen necrosis virus (SNV) (Pulsinelli and Temin 1994). In situations where the 3′end of +sssDNA and the minus-strand DNA copy of the PBS were not complementary, efficient extension of as many as three base mismatches was observed and both wild-type and mutant sequences were found in the PBS region of progeny DNAs, although evidence of rare premature second strand transfer was also found. However, it would appear in the yeast retrotransposon Ty1 that the second strand transfer can occur before all of the tRNA complementary to the PBS is copied (Lauermann and Boeke 1994).
Studies in which targeted deletions were made in the PBS of HIV-1 resulted in deletions and insertions 3′to the PBS in progeny DNAs, supporting the notion that the second jump involves pairing of the plus-strand DNA PBS and its complement in minus-strand DNA (Rhim et al. 1991).
Completion of Integration-competent DNA
Once the second jump has occurred, elongation of the plus and minus strands can continue. When RT extends the minus strand on the plus-strand template, the minus-strand DNA from which the plus-strand was copied must be displaced. RT can carry out displacement synthesis in vitro under appropriate conditions (Boone and Skalka 1981a,b; Huber et al. 1989; Hottiger et al. 1994; Whiting and Champoux 1994), and it seems reasonable to assume that RT (perhaps in concert with NC) carries out this displacement reaction in vivo. The DNA copy of the viral genome is completed when RT copies the plus and minus strands entirely. The final product is a blunt-ended linear duplex DNA (Brown et al. 1989; Roth et al. 1989). This linear product can have a variety of different fates: normal integration, aberrant integrase-mediated circularization (known as autointegration), or joining of the ends by a host ligation activity, forming one circular DNA product with one LTR and another circular DNA product with two LTRs. It has been proposed that the 1-LTR circles could arise through errors during reverse transcription (Junghans et al. 1982a); however, the data indicate that 1-LTR circles are formed in the nucleus after reverse transcription has been completed, suggesting that the 1-LTR circles are formed by host enzymes that can mediate homologous recombination.
Polarity of Strand Transfer
The drawing in Figure 2 depicts an intramolecular strand transfer event—a transfer from the 5′R of one viral RNA to the 3′R of the same RNA—but, at least in theory, it is equally probable that the first strand transfer might be an interstrand event. Whether strand transfer is either intramolecular or intermolecular is referred to as the “polarity” of strand transfer.
If virions are prepared that contain two genetically distinct RNAs that differ in their U3 and U5 sequences, the polarity of the strand transfer can be deduced by examining progeny DNA. During the past several years, three groups of researchers have reported conflicting results regarding the polarity of the first strand transfer (Panganiban and Fiore 1988; Hu and Temin 1990b; Jones et al. 1994). The first group observed that the first strand transfer was always intermolecular (Panganiban and Fiore 1988), the second group found approximately equivalent amounts of intra- and intermolecular first strand transfers (Hu and Temin 1990b), and the most recent study concluded that the first strand transfer is essentially always intramolecular (Jones et al. 1994).
It is not yet clear which result is correct; however, it has been suggested that different rules may apply if the RNA genomes in the virions are intact or broken. In this model, it has been suggested that if the RNA genome is intact, it has a defined structure in the virion, and the first strand transfer is intramolecular; if the RNA is broken, the polarity is random (Jones et al. 1993). It is generally agreed that the polarity of the second transfer is almost always intramolecular (Hu and Temin 1990b; Jones et al. 1993), which presumably reflects the synthesis of only a single duplex DNA in most virions.
- Reverse Transcription of the Viral Genome in vivo - RetrovirusesReverse Transcription of the Viral Genome in vivo - Retroviruses
Your browsing activity is empty.
Activity recording is turned off.
See more...