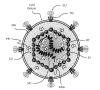
Retroviruses comprise a large and diverse family of enveloped RNA viruses defined by common taxonomic denominators that include structure, composition, and replicative properties (Coffin 1992a,b, 1996). The virions are 80–100 nm in diameter, and their outer lipid envelope incorporates and displays the viral glycoproteins (Fig. 1). The shape and location of the internal protein core are characteristic for various genera of the family. The virion RNA is 7–12 kb in size, and it is linear, single-stranded, nonsegmented, and of positive polarity. The hallmark of the family is its replicative strategy which includes as essential steps reverse transcription of the virion RNA into linear double-stranded DNA and the subsequent integration of this DNA into the genome of the cell.
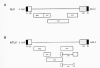
Retroviruses are broadly divided into two categories—simple and complex—distinguishable by the organization of their genomes (Fig. 2) (Coffin 1992a; Murphy et al. 1994). All retroviruses contain three major coding domains with information for virion proteins: gag, which directs the synthesis of internal virion proteins that form the matrix, the capsid, and the nucleoprotein structures; pol, which contains the information for the reverse transcriptase and integrase enzymes; and env, from which are derived the surface and transmembrane components of the viral envelope protein. An additional, smaller, coding domain present in all retroviruses is pro, which encodes the virion protease. Simple retroviruses usually carry only this elementary information, whereas complex retroviruses code for additional regulatory nonvirion proteins derived from multiply spliced messages (Fig. 2). Retroviruses are further subdivided into seven groups defined by evolutionary relatedness, each with the taxonomic rank of genus (Table 1). Five of these groups represent retroviruses with oncogenic potential (formerly referred to as oncoviruses), and the other two groups are the lentiviruses and the spumaviruses. All oncogenic members except the human T-cell leukemia virus–bovine leukemia virus (HTLV-BLV) genus are simple retroviruses. HTLV-BLV and the lentiviruses and spumaviruses are complex.
Table 1
Classification of Retroviruses.
Oncogenic retroviruses occur in all classes of vertebrates, and many act as natural carcinogens. Some of the best studied are Rous sarcoma virus (RSV), a highly pathogenic agent inducing connective tissue tumors in chickens; mouse mammary tumor and murine leukemia viruses (MMTV and MLV); and more recently HTLV (Table 2). The lentiviruses are also found ubiquitously, causing disease principally by killing or inducing loss of function of specific cells and tissues. A representative example is human immunodeficiency virus (HIV), the causative agent of acquired immunodeficiency syndrome (AIDS). Relatively less is known about spumaviruses, which cause no known disease, and interest in this category is relatively recent. The prototypical species is human foamy virus, a virus that has not been definitely linked to a specific pathology.
Retroviruses as Models and Tools
The study of retroviruses has had a broad impact on diverse areas of biology and medicine, notably on molecular genetics, on the study of cellular growth control and carcinogenesis, and on biotechnology (Varmus 1988; Temin 1992). The effect of retrovirology on our concept of genetic information, its molecular forms, transmission, and evolution has been nothing short of revolutionary. In the early years of molecular biology, no exceptions to the unidirectional and presumed irreversible flow of genetic information from DNA to RNA to protein were known. This unidirectional flow came to be known as the “Central Dogma.” It was this Central Dogma that had to be revised when the replication of retroviruses was understood: The retrovirus growth cycle includes as an essential step the copying of RNA into DNA by a virus-coded polymerase, reversing the flow of genetic information, hence the terms “retroviruses” and “reverse transcriptase” (Fig. 3). The reverse transcription of retroviruses is not a singular, odd exception but rather a paradigm for a process that is shared by viral and nonviral genetic elements occurring widely in nature. Examples are the Ty elements of yeast and the copia and ulysses elements of Drosophila (Boeke and Corces 1989; Boeke and Chapman 1991; Evgen'ev et al. 1992; Garfinkel 1992). They resemble retroviruses not only by encoding reverse transcriptase, but also in the structure of both coding and noncoding regulatory sequences of their DNA forms. Indeed, these elements can be viewed as degenerate viruses, although they are more commonly considered to be ancestral to or parallel with Retroviruses in evolution. Among other viral groups, the hepadnaviruses (hepatitis B virus) and the plant-pathogenic caulimoviruses (Shepherd 1989; Robinson 1990), although mechanistically and structurally distinct, practice reverse transcription and are referred to as pararetroviruses. Furthermore, a significant percentage of the mammalian genome appears to be the product of reverse transcription, containing sequences whose characteristics point to RNA as a template precursor. These sequences testify to important evolutionary processes that have shaped the store of vertebrate genetic information but that are not fully understood.
The study of retroviral oncogenesis opened up the entire field of cellular growth control (Bishop 1983; Varmus 1984). It has led to the discovery of Protooncogenes, cellular genes whose products normally function in the transduction of signals that regulate cell replication. Current knowledge of the control of growth and differentiation and of the aberrant growth of cancer has largely originated from investigations of regulatory genes first identified in highly oncogenic retroviruses. Studies of these genes have highlighted the importance of discrete genetic changes in malignant transformation.
Retroviruses have also provided essential tools for biotechnology. Reverse transcriptases from avian and murine retroviruses are used universally to generate cDNA copies of RNA which then can be manipulated with relative ease for cloning and sequencing (Skalka and Goff 1993). Modified retroviral genomes are widely used in transient and stable expression of cloned genes in vertebrate cells (Miller 1992). They are also promising delivery vehicles for human gene therapy.
A Unique Genetic Strategy
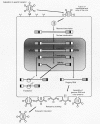
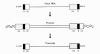
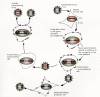
The retroviral replication cycle follows the general pattern of enveloped viral infections, but embedded in it are some highly uncommon features (Swanstrom and Vogt 1990; Luciw and Leung 1992; Coffin 1996). Retroviruses enter the host cell through the attachment of their surface glycoproteins to specific plasma membrane receptors, which leads to fusion of virus and cell membranes (Fig. 3). The interaction of virus and cell surfaces is highly specific; it constitutes the main determinant of viral host range, defining susceptible animal species and target cells within the host. After penetration into the cell, the RNA genome, still contained in a core complex of nonglycosylated proteins and associated with the virion reverse transcriptase, is transcribed into a double-stranded DNA. Transcription into DNA involves two jumps of the reverse transcriptase from the 5′terminus to the 3′terminus of the template molecule. The result of these jumps is a duplication of sequences located at the 5′and 3′ends of the virion RNA; these sequences then occur fused in tandem on both ends of the viral DNA, forming the long terminal repeats (LTRs) (Fig. 4). The LTRs regulate viral gene expression and therefore replication and pathogenesis.
Reverse transcription takes place in the cytoplasm; the viral DNA is translocated into the nucleus where the linear copy of the retroviral genome is inserted into chromosomal DNA with the aid of the virion integrase to form a stable provirus. Integration does not permute the linear order of the proviral sequences, LTR-gag-pol-env-LTR (Fig. 4). The number of possible sites of integration into the cellular genome is very large and very widely distributed. With integration, the provirus achieves the status of a cellular gene and is expressed through the agency of cellular RNA polymerase II and replicated by cellular enzymes in concert with chromosomal DNA. Control of proviral transcription remains largely with the noncoding sequences of the viral LTR. Transcription of the provirus generates spliced and unspliced mRNAs and full-length progeny RNA genomes. In infection with simple retroviruses, control of transcription is mediated solely by interaction of cellular factors with the DNA of the LTR. Complex viruses, in contrast, take a more active role, encoding trans-activating factors that affect the levels of transcripts and the relative amounts of the products of the various genes. This strategy allows these viruses a measure of control of gene expression not seen with the simple viruses.
Viral messages are translated on cellular ribosomes. The translation products, together with progeny RNA, are assembled at the cell periphery into viral particles that are released from the cell by budding of the plasma membrane. Budding of viruses is followed by proteolytic cleavage of virion polyproteins by a viral protease and by cellular proteases. Productive Retroviral infection is not of necessity cytopathic; infected cell cultures often show no visible effects of viral production. In some congenital infections in animals, virus can be produced by most cells in most tissues without deleterious effect on the development and function of the organism (Rubin et al. 1961, 1962).
In the replication cycle, viral entry, assembly, and release follow common patterns (pathways) similar to these events in other enveloped RNA viruses. The unique steps in the retroviral growth cycle are reverse transcription and, especially, integration. Reverse transcription generates a progenitor proviral DNA copy from which the entire viral progeny of the cell is derived by polymerase-II-mediated transcription. Although reverse transcription is much more error prone than cellular DNA replication, its product, once integrated into the cellular genome, has the genetic stability of cellular genes. Reverse transcription and integration make retroviral infection permanent, as integrated proviruses are only rarely lost from the cellular genome: “A retrovirus is forever.” Integration of viral DNA into the host genome is ipso facto mutagenic. This insertional mutagenesis can inactivate or activate cellular genes; this process is one of the Mechanisms by which retroviruses induce tumors.
Viral Takeover at the Level of the Gene
Retroviruses have also developed unique mechanisms of pathogenicity involving the transfer or transcriptional activation of specific cellular genes. These Mechanisms are based on genetic recombination between virus and cell and between viral genomes. Retroviral particles contain two copies of their genome linked by regions near the 5′termini. They are diploid and are the only viruses so equipped (Billeter et al. 1974; Kung et al. 1975; Beemon et al. 1976). A direct consequence of diploidy is the formation of heterozygote virions in cells that are infected with two or more genetically distinct but related retroviruses. Such heterozygotes give rise, in the next cycle of infection, to stable genetic recombinants that are formed during the process of reverse transcription of the two parental genomes from the same viral particle. Rates of recombination between related retroviruses are high (Coffin 1979; Linial and Blair 1982; Temin 1991).
Interviral genetic exchange, together with integration into the cellular genome, formally also a recombination event, probably accounts for the presence of cellular genes in some retroviruses (Fig. 5). A plausible hypothesis for this acquisition of cellular genetic material postulates that a provirus integrates upstream of a cellular gene and leads to the production of chimeric virus-cell transcripts. In the next round of replication, nonhomologous recombination between virus and cell sequences leads to the incorporation of the cellular gene into the retroviral genome, so that it is now transported by the virus from cell to cell and expressed under control of the viral LTR (Goldfarb and Weinberg 1981; Swanstrom et al. 1983; Raines et al. 1988; Felder et al. 1991, 1993; Coffin 1992b; Swain and Coffin 1992; see Chapter 4). The usual product of this transduction process has acquired the host sequence at the cost of one or more viral genes. Such viruses are therefore generally defective for replication, requiring the presence of a replication-competent provirus in the same cell to provide viral proteins for replication (see Fig. 4). The transduction of cellular genes has been found only with simple retroviruses and not with complex retroviruses. The reasons for this difference are not clear, but they may have to do with the mechanism by which retroviruses acquire cellular sequences or with viral genome organization that must be tolerant of foreign inserts.
The modified cellular genes carried by retroviruses convey a high degree of tumorigenicity to the virus. These viral or v-onc genes are usually mutated growth-regulatory genes. Their cellular progenitors are referred to as Protooncogenes or c-onc genes (Bishop 1983; Varmus 1984; Cooper 1990). Overexpression or inappropriate expression, often combined with mutation of an oncogene that has become part of a viral genome, results in a gain of function of a positive growth signal. This constitutive gain of function induces and maintains malignant transformation. Retroviruses with oncogenes in their genomes are particularly fast-acting carcinogens and in most cases also transform cells in culture. Retroviruses lacking an oncogene do not transform cultured cells, but some can induce tumors in animals, a process that is characterized by a long latent period. Here again, an oncogene is the mediator of neoplastic transformation, but in this instance, it is the cellular homolog that is activated through the insertion of a provirus. The viral LTR through its promoter and enhancer sequences alters expression of the neighboring cellular oncogene (for reviews, see Lazo and Tsichlis 1990; van Lohuizen and Berns 1990; Kung and Vogt 1991). Transduction and insertional activation of cellular oncogenes are the two main mechanisms by which most retroviruses induce tumors. Other mechanisms exist but are less common. One is exemplified by the highly aggressive leukemias which are linked to infection with HTLV. This virus neither carries a transduced oncogene in its genome nor activates a resident oncogene by insertion; it may cause tumors through one of its regulatory proteins, possibly by changing the expression levels of cellular genes (Feuer and Chen 1992). A unique mode of tumor induction is also seen with a form of the Friend murine leukemia virus. It produces a modified Env protein that appears to mimic a component of the erythropoietin signaling pathway (Li et al. 1990; D'Andrea et al. 1992; Johnson and Benchimol 1992).
Retroviruses show the entire continuous spectrum of effects on cell replication and survival from pronounced growth stimulation to cell death. The latter is an important element in lentiviral infections including infection with HIV. Although it appears likely that viral Env proteins are involved in some aspects of retroviral cell killing, as is reinfection of cells leading to accumulation of large amounts of viral DNA, the full cytopathic mechanisms operating at the cellular and organismic levels remain to be established.
Publication Details
Copyright
Publisher
Cold Spring Harbor Laboratory Press, Cold Spring Harbor (NY)
NLM Citation
Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 1997. The Place of Retroviruses in Biology.