MicroBIGG-E Map Details
This is an alpha release. For feedback, send an email to: pd-hep@ncbi.nlm.nih.gov
This data resource reflects assembled genome sequence data for bacterial pathogens in GenBank with acquired resistance genes and point mutations identified by AMRFinderPlus and with geographic location metadata. The presence/absence of selected gene families and individual genes and alleles, with respect to geography, needs to be carefully interpreted in the context of the sequence data generated from each country. These sequences might not reflect global patterns of phenotypic resistance. Due to differences in collection and decisions to sequence genomes, within- and cross-country data should be interpreted with caution.
Dataflow diagram for understanding the context of the sequences show in the MicroBIGG-E map.
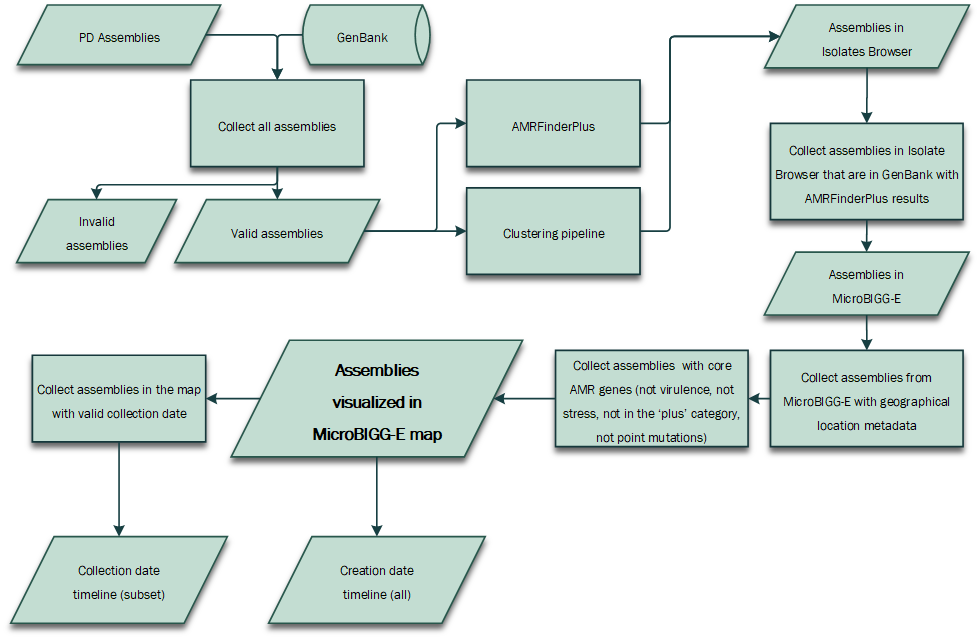
- For each organism group, the assemblies produced by the Pathogen Detection pipeline , and those already in GenBank, are collected. Only valid assemblies are included. Invalid assemblies include those flagged as anomalous in the Assembly archive, and those that fail validation in the Pathogen Detection pipeline.
- For inclusion in the Isolates Browser, all assemblies for a given organism group are clustered. The assemblies in the Isolates Browser include those that cluster together, as well as singletons. Antimicrobial resistance genotypes identified by AMRFinderPlus are included if the results are available at the time of publication. Each organism group is indepedently published.
- Valid assemblies in GenBank with AMRFinderPlus results are included in the MicroBIGG-E. Assemblies that are included in the Isolates Browser, but have a validation issue preventing submission to GenBank (ex: missing strain information), or that do not have AMRFinderPlus results (either no genes/proteins identified, OR the results are not available yet) are not included.
- Those assemblies in MicroBIGG-E that have valid geographic information in the "geo_loc_name” field are in scope for the MicroBIGG-E Map and that have acquired resistance genes (those in the ‘core’ category). The numbers in parentheses in the organism group selector reflect this denominator of isolates. Point mutations, AMR genes in the ‘plus’ category, virulence genes, and stress response genes are not currently included. If you are interested in these additional categories, email us at: pd-help@ncbi.nlm.nih.gov.
- Every isolate that is assembled by the Pathogen Detection pipeline has a "create_date" timestamp for when the data entered the system. Only those where the submitter supplied a valid "collection_date" metadata item are included in the Collection Date timeline.
- Note these counts include partial sequences (Method: PARTIAL and PARTIAL_END_OF_CONTIG) and sequences with internal stops (Method: INTERNAL_STOP).
