NCBI Genome Data Viewer
The NCBI Genome Data Viewer (GDV) is a genome browser supporting the exploration and analysis of annotated eukaryotic genome assemblies. The GDV browser displays biological information mapped to a genome, including gene annotation, variation data, BLAST alignments, and experimental study data from the NCBI GEO and dbGaP databases. GDV release notes describe new features relating to this browser.
The core component of the GDV browser is the NCBI Sequence Viewer, which supports analysis of genomic assemblies at multiple levels, from the whole chromosome or scaffold to the sequence base pair. Please refer to Sequence Viewer documentation and release notes for more information.
Go to the GDV YouTube playlist to view video tutorials that can help you better use this application in your work. Send us feedback if you have any questions.
- Finding organisms and genome assemblies
- Using the browser
- Page widgets
- Controlling GDV with URL Parameters
- Cite Us
Finding Organisms and Genome Assemblies
The GDV home page (Figure 1A) allows you find and select organisms and assemblies available to view in the browser. You can search directly for common or scientific names using the search box, or click on nodes in the tree to explore different taxa. Hovering over tree nodes with '+' signs will open a tooltip that reveals the number of additional nodes with assembly data. Clicking on these nodes will zoom the tree to the next deeper level. To return to the top level tree after zooming in, click the '<<' icon. To go up one taxonomic rank instead, click the '<' icon. Clicking on any of the organism-specific nodes will update the information panel on the right side of the page with information pertaining to the assembly or assemblies that is available in the browser.

Figure 1A: Default (tree) view of the GDV home page used to browse and search genomes and select assemblies to view in the genome browser.
Enter gene names, dbSNP ids, phenotypes, assembly components/scaffolds, or sequence accessions into the search box on the assembly information panel. Examples of searches relevant to the selected organism are shown below the box to assist you in constructing queries. You can provide location information as a range, point, or cytogenetic band. If your search has an exact location match, you will be taken directly to that location in the genome browser; otherwise, you will be shown a search results panel. For more information on browser searches, see the Search section of this document.
If you don't wish to perform a genome search, use the Browse genome button to go to a default location, typically the whole molecule view of the first chromosome for chromosome-level assemblies or the longest scaffold for scaffold-level assemblies. You can also click on a chromosome ideogram at the bottom of the panel to go to the corresponding chromosome in the genome browser. Other options in this table will take you outside of the Genome Data Viewer to related analysis resources at NCBI, including the Comparative Genome Viewer (CGV), BLAST, and the NCBI Datasets genome table. You can click on the (i) icon to obtain the annotation report for NCBI-annotated assemblies.
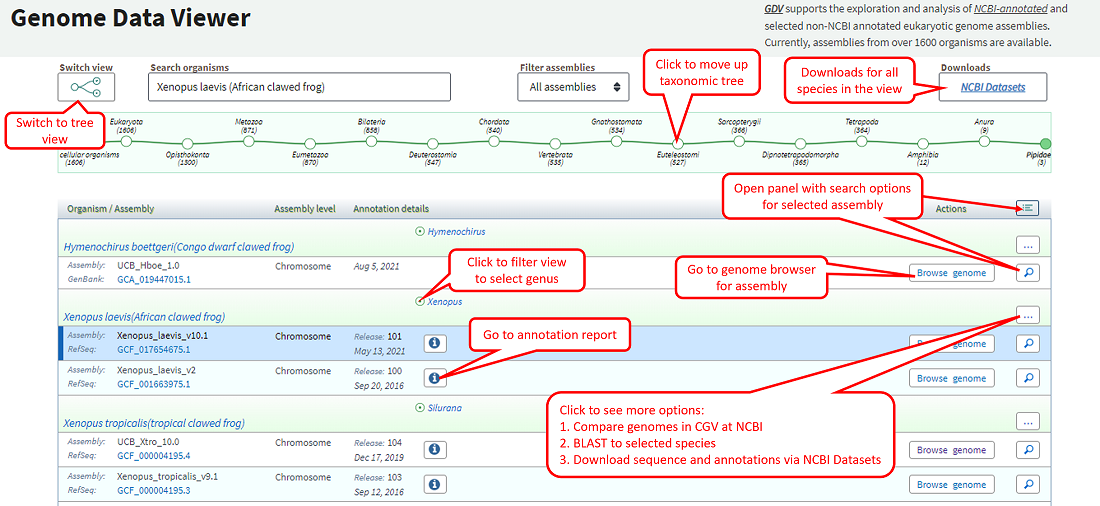
Figure 1B: Table view of the GDV home page used to browse and select assemblies.
The GDV home page also offers an alternate table configuration (Figure 1B). Here, you can browse through a table of all the assemblies in a taxonomic grouping sorted by species. This view may be helpful for selecting an assembly of interest based on assembly name, assembly level (scaffold vs chromosome), annotation release number, or annotation date. You can use the Filter assemblies option to limit the organisms and assemblies displayed in the table to RefSeq accessions that contain annotation from NCBI's annotation pipeline.
The table view contains options to go to the NCBI Comparative Genome Viewer (CGV), BLAST, or NCBI Datasets genome table for a selected organism. You can also obtain the annotation report or go to the genome browser view for a selected assembly of interest. Clicking on the magnifying glass icon opens a details panel (Figure 1C) with additional options, including the option to search within a selected genome assembly or navigate to a chromosome in a chromosome-level assembly.
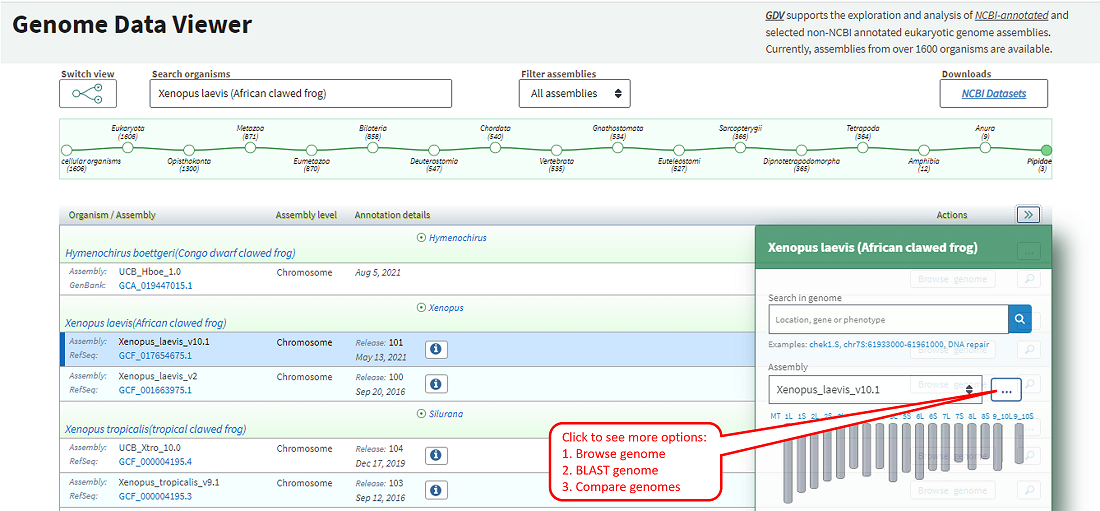
Figure 1C: Table view showing details panel for a selected assembly in the table.
The horizonal tree diagram above the table allows you to browse up the taxonomic tree. On the upper right, there's a link to NCBI Datasets to download genomic sequence and annotation from any or all organisms in the current table view. You can return to the tree view from this page at any time via the button on the upper left of the page.
Using the Browser
GDV is comprised of a series of page elements (widgets) that are used for different types of interactions with the browser, such as genome searches, analysis of BLAST results, data uploads, or changing the display. The widgets communicate with one another such that an action in one widget causes other widgets on the page to update. For instance, clicking on a chromosome in the Ideogram View will update all other widgets on the page to show information relevant to that chromosome.
An announcement banner may appear along the top of the browser page above all the widgets. Clicking the x on the right end of the banner will remove it from the GDV session. The banner will reappear when the user clicks "Reset All", arrives at a fresh session of GDV, or when the banner message has been updated.
Page layout
Links
The following links are available in the upper right of the browser header:
- Home: Go to the GDV landing page. Clicking on the "Genome Data Viewer" text on the upper left will also direct you to the landing page.
- Share this page: Generate a temporary URL (90 days) to GDV with current track and display settings
- Reset All: Reset all settings in the browser to default settings
- More Tools: Additional genome analysis resources at NCBI
- Compare genomes using the Comparative Genome Viewer (CGV)
- Download genomes from NCBI Datasets
- BLAST this genome
- More Info: Menu with additional options
- Help: GDV documentation
- FAQ
- Release notes
- Feedback
- Browser agreement : Documentation describing agreement among NCBI, UCSC and Ensembl regarding assemblies that may be shown in genome browsers
Page Overview
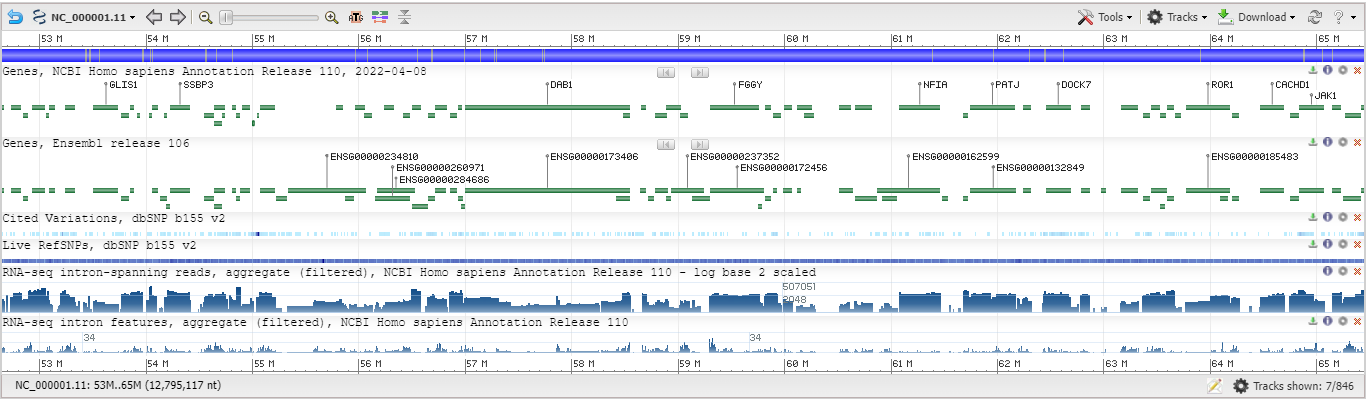
Below is an overview of GDV that highlights each of the main page elements (Figure 2A). The left sidebar contains a series of widgets that provide tools that can be used to manupulate the display. The center of the page contains an instance of the NCBI Sequence Viewer where tracks and track data is visualized.
This GDV browser view can be customized in several ways. The caret arrow can be used to show or minimize each of the widgets on the left sidebar. These widgets can also be re-ordered by pressing within the header to select, and then dragging and dropping the widget into a desired location in the sidebar.
The entire left sidebar can be hidden by clicking on the blue << icon or by pressing "[" on the keyboard (Figure 2B and Figure 2C). Clicking the >> icon or pressing "[" again will reveal the left sidebar options once again.
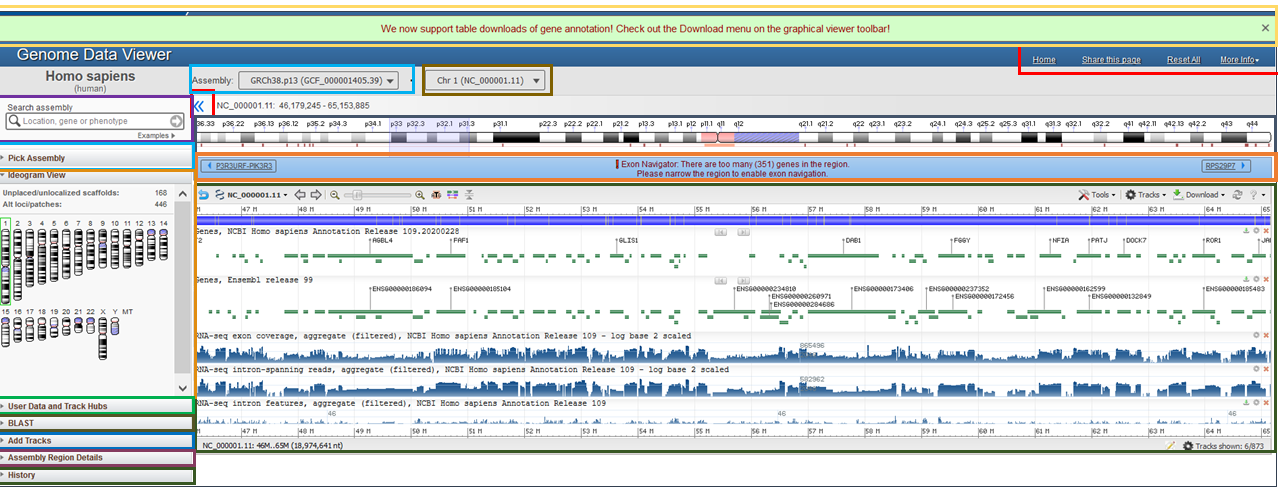
Figure 2A: A page overview with individual page elements highlighted in different colors.
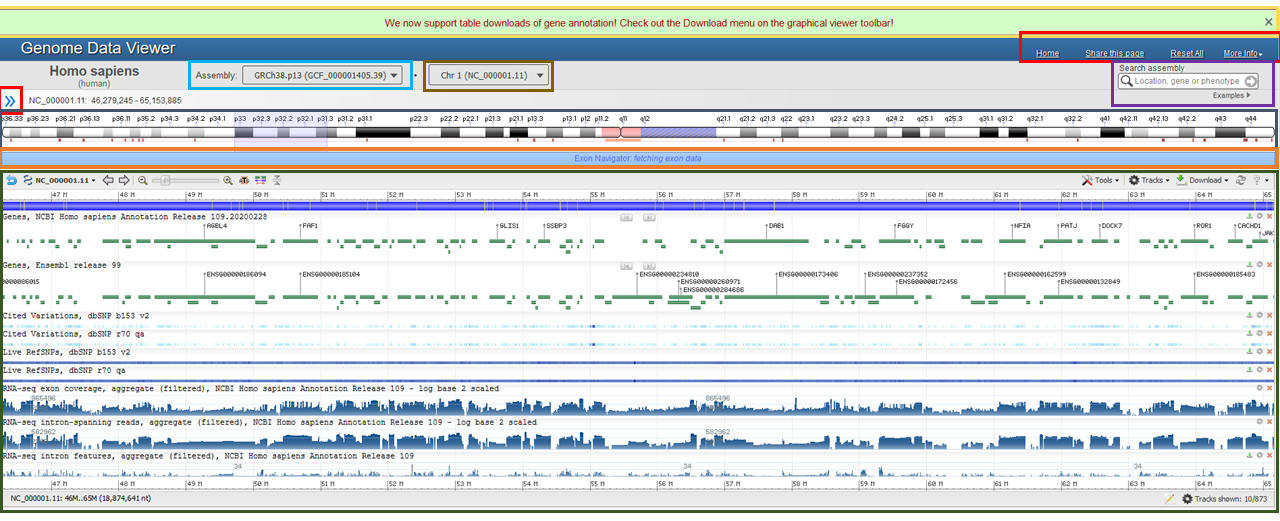
Figure 2B: A page overview with the left sidebar hidden and page elements highlighted. Note that the search box is in a different position on the upper right.

Figure 2C: Switch Button to hide/reveal the left sidebar.
Page Widgets
Below is an overview description of the principal widgets or elements on the page.
Assembly and Chromosome
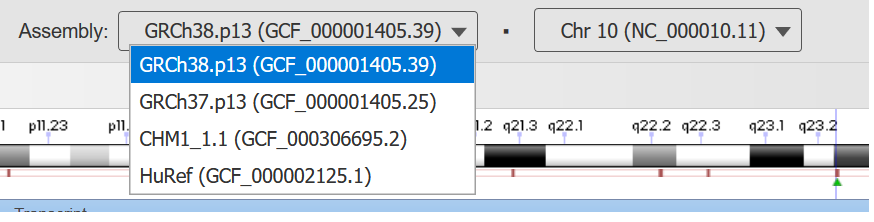
Figure 3. Assembly and chromosome indicator/selector menu on the top of the browser page.
The Assembly and Chromosome that are viewed in the genome browser are indicated on the top center of the page (Figure 3). The current sequence accession and coordinates appear on the line below.
The Assembly Indicator appears as a dropdown menu when there are multiple assemblies available in GDV. You can use this menu to switch to view a different assembly for the selected organism.
The Chromosome Indicator appears as a drop-down menu that allows you to navigate to different chromosomes or scaffolds within the assembly. For chromosome-level assemblies, the chromosomes in the assembly will be listed in the menu. For assemblies that lack assembled nuclear chromosomes, (e.g. https://www.ncbi.nlm.nih.gov//genome/gdv/browser/genome/?id=GCF_000297895.1), the menu will contain up to the largest 30 scaffolds/contigs by size. The "More..." option at the bottom of the menu links to the NCBI Nucleotide search results page listing all scaffolds for the given assembly.
Chromosomes or scaffolds that have been viewed in a genome browser session (including scaffolds not in the top 30) will be listed in the selection menu under a separate sub-heading for ease of future access. These added scaffolds will disappear from the menu when you click "Reset All" on the upper right or start a new genome browser session.
Search
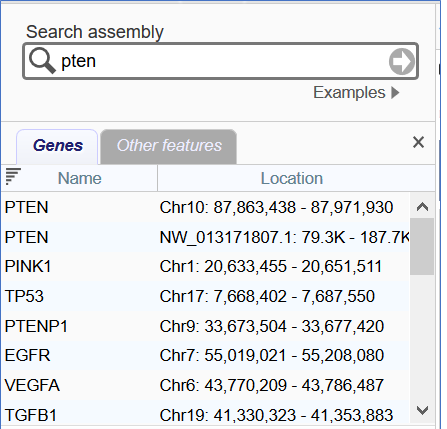
Figure 4. Search box with search results shown below. This box appears on the upper right of the page when the left sidebar is hidden.
The Search Box (Figure 4) is located on the upper left of the page by default and can be used to find a gene or location within the selected assembly. You can enter a location, a gene name, SNP RSID, HGVS pattern, Ensembl gene/transcript ID, phenotype, or disease term (such as 'PTEN' or 'rs13432' or 'diabetes'). Location information can be provided using a coordinate range or a single point. This search box can also be used to search and navigate to a scaffold or contig within the assembly (e.g. NW_011934491.1). If your search finds an exact and unique match, the Sequence Viewer and other widgets on the page will update to show the location of that match. If you perform a search without a unique match, a new panel will appear with a list of results and their locations. The Ideogram View and Chromosome Region Selector will also be updated to display clickable colored arrowheads indicating the location of search results.
In the search results panel, the Genes tab lists genes matching the search term, while the Other Features tab lists related transcripts or other features. When you click on a row in either tab of the search results, or on the arrowheads in the Ideogram View or Chromosome Region Selector, the Sequence Viewer and other page elements will update to go to that location. You can click on the x to hide the search results panel. Recent search terms will be remembered by default and shown in a pop-up menu when the cursor is placed within the search box.
When the left sidebar is hidden, the search box appears in the upper right of the page. The right search box has identical behavior to the default search box, and recent searches made in both locations can be retrieved from either location.
Pick Assembly
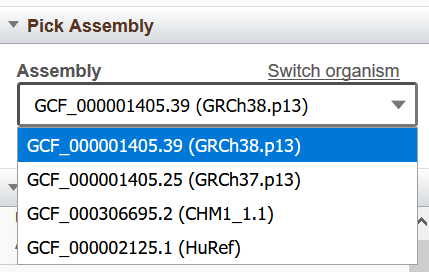
Figure 5. Pick Assembly Widget.
The Pick Assembly widget (Figure 5) indicates the current genome assembly and annotation context for GDV. This widget appears in the left sidebar when there are multiple assemblies available for an organism. It provides a drop-down menu of available assemblies. Click on the "Switch organism" link to go to the Genome Data Viewer landing page to select from the full list of available organisms and assemblies.
Ideogram View
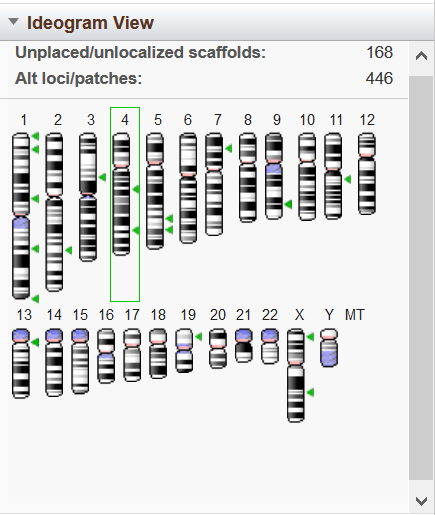
Figure 6. Ideogram View. Green arrowheads indicate locations of genes found in the most recent search in GDV.
The Ideogram View shows the chromosomes in a NCBI-annotated chromosome-based assembly (Figure 6). A green highlight surrounds the current chromosome that is viewed in the display. Clicking on a chromosome will update the Sequence Viewer to the selected chromosome. No chromosome ideograms will be displayed for scaffold- or contig-based assemblies.
The locations of gene(s) or SNP found in the most recent GDV search appear as green or blue arrowheads, respectively, on the chromosomes in this widget. When you hover over a arrowhead, a panel appears with more information. If data is loaded to the BLAST widget, BLAST hits on the assembly will be shown as orange arrowheads. You can click on an arrowhead or within a pop-up panel to navigate to the corresponding location in the Sequence Viewer display.
The Ideogram View widget also provides a count of the non-chromosomal sequences in the assembly. Separate counts are provided for unlocalized/unplaced scaffolds and for alternate loci/patch scaffolds. The View Scaffolds link goes to the NCBI Nucleotide search results page listing all non-chromosome scaffolds. For more information on these types of scaffold sequences, please see the documentation for the NCBI Assembly Model.
Chromosome Region Selector

Figure 7A. Chromosome Region Selector widget.
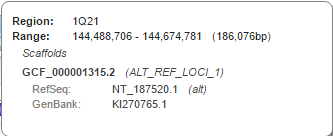
Figure 7B. Region pop-up information panel.
The Chromosome Region Selector (Figure 7A) is an interactive ideogram that provides context and navigation for the genome browser. The ideogram corresponds to the current chromosome viewed in browser, and the boxed area in blue denotes the region currently shown in the Sequence Viewer. The Chromosome Region Selector widget will automatically update if you navigate or zoom using the Sequence Viewer or other elements on the page.
You can use the Chromosome Region Selector to navigate to a different region in the Sequence Viewer by clicking and dragging within the ideogram to select a different boxed window. You can also select the ends of the blue box with your cursor and adjust the size of the region. Clicking within a chromosome band (if applicable) will create a box around that band and navigate the Sequence Viewer directly to the sequence corresponding to that cytological region.
The locations of search results (gene or SNP), markers set in the Sequence Viewer, or BLAST results from the BLAST widget will appear as colored arrowheads (gene = green, SNP or marker = blue, BLAST = orange) on the ideogram. When you hover over an arrowhead, a panel appears with more information. You can click on an arrowhead or within a pop-up panel to navigate to the corresponding location in the Sequence Viewer display.
A thin line beneath the ideogram shows regions of the chromosome for which there are alternate loci or patch scaffold sequence representations generated by the Genome Research Consortium (GRC). Hovering over any of the red-colored portions of the line will open a region-specific information panel (Figure 7B) containing the region name and coordinates, as well as sequence identifiers for associated alternate loci or patches. Clicking on the region name or RefSeq accession will take you the corresponding region or accession in the Sequence Viewer, while clicking on the GenBank accession will open the GenBank record in the NCBI Nucleotide database. More information about GRC assembly regions can also be found in the Assembly Region Details widget (Figure 7C), which is located by default toward the bottom of the left sidebar.
Refer to this video for a demonstration of how to use the Chromosome Region Selector.
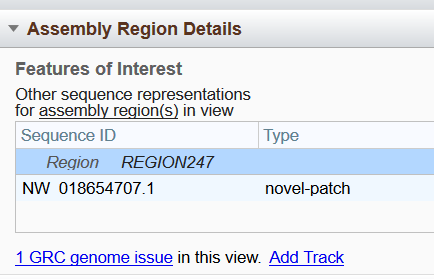
Figure 7C. Assembly Region Details widget.
Exon Navigator

Figure 8a. Exon navigator.
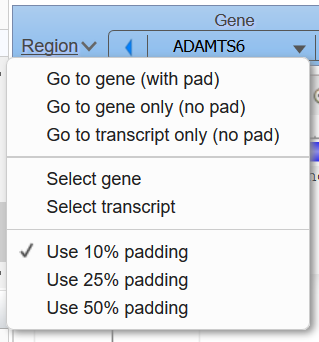
Figure 8b. Region menu in the Exon navigator contains options for resetting the view around the selected gene.
The Exon Navigator element (Figure 8a) appears as a blue bar below the Chromosome Region Selector widget. This widget contains drop-down selection menus and arrows that allow a user to navigate among neighboring genes, transcripts of a gene, and exons of a transcript. The Region link opens a menu (Figure 8b) containing options to configure the padding of the display around the selected gene or trancript. These options are not available when the view is zoomed further out; instead, a warning message appears listing the number of genes annotated on the latest NCBI Genes annotation release in the current display window.
When a single gene is selected, open circles appear toward the right of the Exon Navigator corresponding to the exons of the selected transcript of the selected gene. You can navigate the Sequence Viewer to a particular exon by clicking on a circle in this widget or using the left/right arrows to jump to upsteam or downstream exons. Hovering over an exon circle reveals text showing the exon number (relative to the selected transcript) and coordinates on the chromosome accession. Right-clicking on an exon circle reveals an option to view a pop-up window showing the genomic sequence of the exon in FASTA format. Users can copy text from this window for their own use.
Sequence Viewer
Figure 9. NCBI Sequence Viewer.
The NCBI Sequence Viewer (Figure 9) is an application that provides a linear graphical representation of features annotated or aligned to individual sequence accessions. You can use the pan arrows and zoom slider on the left side of the Sequence Viewer toolbar to navigate within this viewer; changes in the displayed range will be automatically propagated elsewhere in the genome browser, including to the Chromosome Region Selector and the Exon Navigator.
You can access NCBI Primer-BLAST, set markers, download sequence and track data, generate a PDF or SVG image, and more from within the Sequence Viewer toolbar and context menus. The Tracks button (in the upper right of the toolbar) allows you to configure the display with tracks provided by NCBI or to add custom tracks and BLAST request IDs.
Additional information on using the Sequence Viewer (which is embedded on many NCBI pages) can be found on the Sequence Viewer documentation home page.
Tracks and User Data
The Tracks and User Data widget (Figure 10) allows you to add additional tracks for display in the Sequence Viewer. You can find NCBI-provided tracks, connect to a track hub, upload files, or stream data from remotely-hosted files. Click on the Options drop-down menu to access the different ways to add tracks.
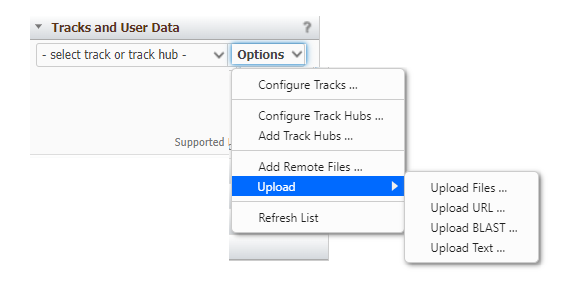
Figure 10. Tracks and User Data widget.
If you are logged into a My NCBI account, your custom tracks and track hubs will be available to you in other NCBI Sequence Viewer instances and browsers displaying the same assembly version (e.g. Variation Viewer). Uploaded tracks will expire 60 days after they are last accessed; streamed tracks and track hubs will persist until you remove them.
NCBI-Provided Tracks
There are often more data tracks available for a given assembly in GDV than are displayed by default. Click on Configure Tracks in the Options menu (Figure 10) to launch the Configure Tracks panel (Figure 11). Here, you can browse and search for additional NCBI-provided tracks to add to your view. You can also access this panel by going to the "Tracks" menu in the Sequence Viewer toolbar and selecting the Configure Tracks option.
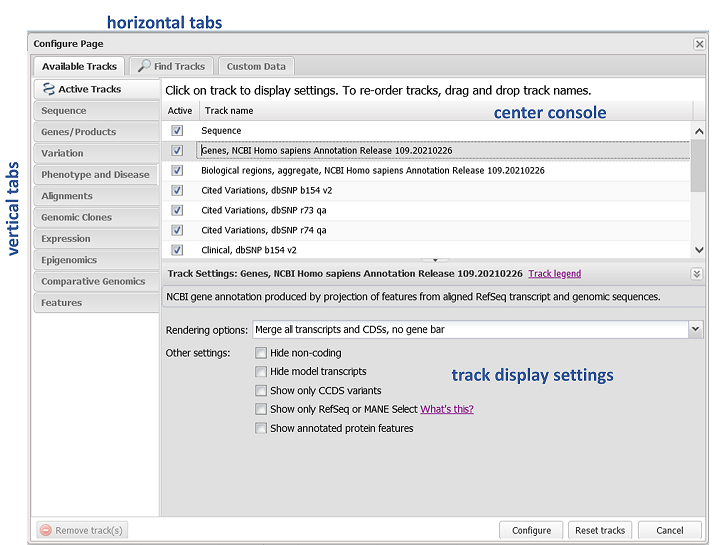
Figure 11. Configure Tracks panel. This panel is also accessible from the Tracks menu on the Sequence Viewer toolbar.
By default, when you first open the Configure Track panel, the Active Tracks group will be open. This is the set of tracks that are currently shown in the display. You can re-order tracks in this group by dragging and dropping the track name in the center console. To remove a track from the display, uncheck the box. Browse the vertical tabs to find additional tracks of interest to add.
Go to the Find Tracks horizontal tab to search by keyword all tracks that are available in GDV for your assembly. You can also search outside of GDV for aligned study data from the NCBI GEO (Gene Expression Omnibus) and SRA (Sequence Read Archive) databases and GWAS data from dbGaP (human reference assemblies only). This search will only return data that is aligned to the assembly currently in view.
To change how a track is displayed, select the track and adjust the options in the track display settings section of the page. Once you are done configuring the display, click on the Configure button in the lower right hand corner to apply your changes.
You can also access display options for particular tracks using the configure gear icon located in the track title tooltip or on the right side of the track.
Upload Files
You can upload files by going to the Upload sub-menu in the Options menu (Figure 10) or by dragging the file into the widget. The following file types are supported: BED, GFF3, GTF, GVF, VCF, HGVS, ASN.1 (text and binary).
A progress bar at the bottom of the widget will indicate the status of the upload or initial connection to a remote data file (Figure 12). If there are sequences in your file that are not part of the assembly displayed in GDV, a red warning icon will appear next to the confirmation message. However, you will still be able to view tracks for sequences in the file that are part of the assembly. Click on the Details link in the status message to view warnings or error messages associated with uploading or streaming your custom track data.
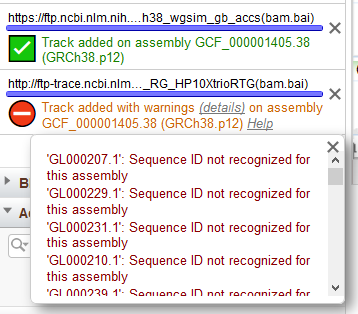
Figure 12. Confirmation messages that appear when a custom track is added. Note that a track is added to the Sequence Viewer display in both cases.
Concurrent upload of multiple files is supported. The upload limit is currently 4 GB per file. The file name will be used as the track's display name unless an alternate name (optional) is provided. Once uploaded, the track will appear in the Sequence Viewer display with a green title background and (U) designation and in the drop-down menu of the User Data and Track Hubs widget (bolded if in the current display). To remove tracks, click on the X to the right of the track name in the drop-down menu. File uploads can also be managed the Configure Tracks panel (Figure 11).
If an uploaded track has discrete features (e.g. SNPs or gene annotations) on the currently displayed sequence, those features will be listed in a paginated table when that track is selected from the drop-down track selection menu (Figure 13). Hovering the mouse over a table row opens a tooltip with feature details. Click on a row to go to the location of that that feature in the Sequence Viewer. Use the menus below the table for additional table navigation.
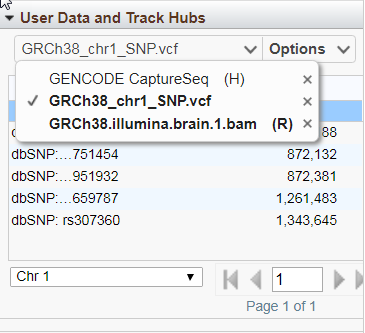
Figure 13. User data feature table.
BLAST Results
You can enter an RID (request ID) from a BLAST search into the Tracks and User Data widget in the Upload sub-menu in the Options menu (Figure 10). The results will appear as an alignment track in the display. BLAST results can also be added via the Custom Data horizontal tab of the Configure Tracks panel (Figure 11). To take advantage of the full range of BLAST analysis tools in GDV, refer to the BLAST widget.
Remote Streaming Files
BAM, indexed VCF, VCF, bigBED, and bigWIG files hosted at publicly-accessible remote locations can be streamed for display in GDV. To add these data as tracks, select Add Remote Files from the Options menu (Figure 10) and enter the corresponding URL in the appropriate box. If viewing a BAM file, an appropriate index file must be available in the same remote location. The file name will be used as the track’s display name unless an alternate name (optional) is provided.
A progress bar at the bottom of the widget will indicate the status of the upload or initial connection to a remote data file (Figure 12). If there are sequences in your file that are not part of the assembly displayed in GDV, a red warning icon will appear next to the confirmation message. However, you will still be able to view tracks for sequences in the file that are part of the assembly. Click on the Details link in the status message to view warnings or error messages associated with uploading or streaming your custom track data.
Once connected, remotely hosted files will appear in the Sequence Viewer with a green title background and (R) designation, and will be listed in the drop-down menu of the Tracks and User Data widget (bolded if in the current display). To remove or disconnect from tracks, click on the X to the right of the track name in the drop-down menu. Remotely hosted files can also be managed via the Configure Tracks panel (Figure 11).
Track Hubs
GDV can display content provided in track hubs format. Select Add Track Hubs from the Options menu to access the Track Hubs panel (Figure 14A). If you’ve already added a track hub(s), you can select Configure Track Hubs to go directly to configure options for your hubs.
See this brief video or this webinar for more information on adding track hubs at NCBI.
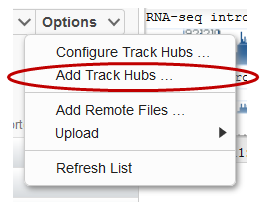
Figure 14A. Getting started with Track Hubs.
If you know the URL of the track hub, you can enter it directly into the dialog in the Track Hubs panel (Figure 14B). Alternatively, you can search or browse the Track Hub Registry by keyword or data type. Select a search result to connect to the corresponding hub (Figure 15).
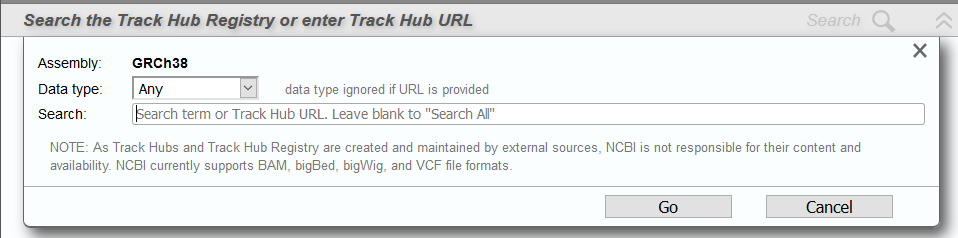
Figure 14B. Search/add Track Hubs dialog.
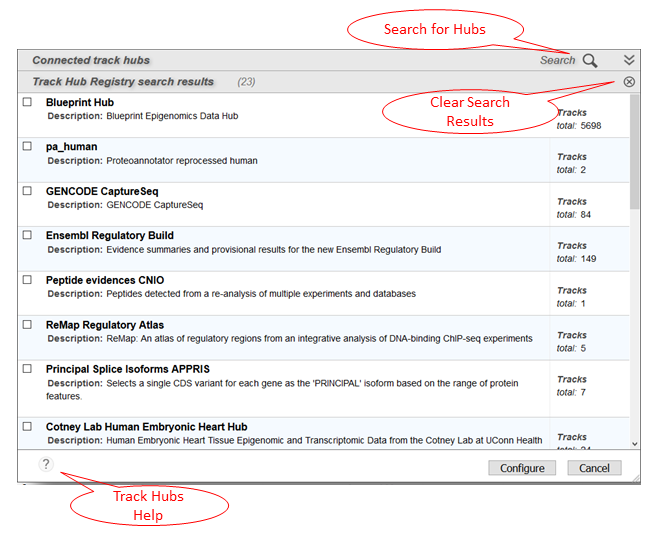
Figure 15. Track Hubs panel showing search results.
A question mark icon on the lower left of the Track Hubs panel shows you additional help documentation for this dialog (Figure 15). You can clear your search results by clicking on the circled x icon on the upper right of the panel.
Upon connecting to a hub, the Tracks and User Data widget will display summary information for the selected hub (Figure 16). Connected hubs will also be listed in the drop-down menu of the widget designated by (H).
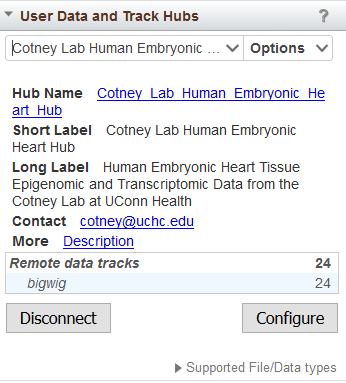
Figure 16. Track Hub summary information in the Tracks and User Data widget.
Connecting to a hub will collapse the search result view (Figure 15) in the Track Hubs panel and expand the configuration view (Figure 17). From this interface, you can select tracks for display, configure track display options, and view hub and track metadata. Currently, you will only be able to connect to tracks that are in BAM, VCF, bigWig/multiWig, or bigBED format.
Tracks that the track hub provider has suggested for default display can be distinguished by their black font, while non-default tracks will be listed in light red font. Check the box next to one or more track names to connect to the track. A green circle will appear to the right of a track name once GDV has connected to a track. All connected tracks will show the green ball icon regardless of whether they are visible in the Sequence Viewer display. If GDV fails to connect to the track, an error message will be shown.
Click on the hamburger icon to the right of track groupings (e.g. composite track, super track) to access the bulk options dialog (Figure 17 inset). This menu allows you to select or unselect all tracks in that grouping that are marked by default by the hub provider. You can also choose to list only default tracks, i.e hide non-default tracks from the menu.
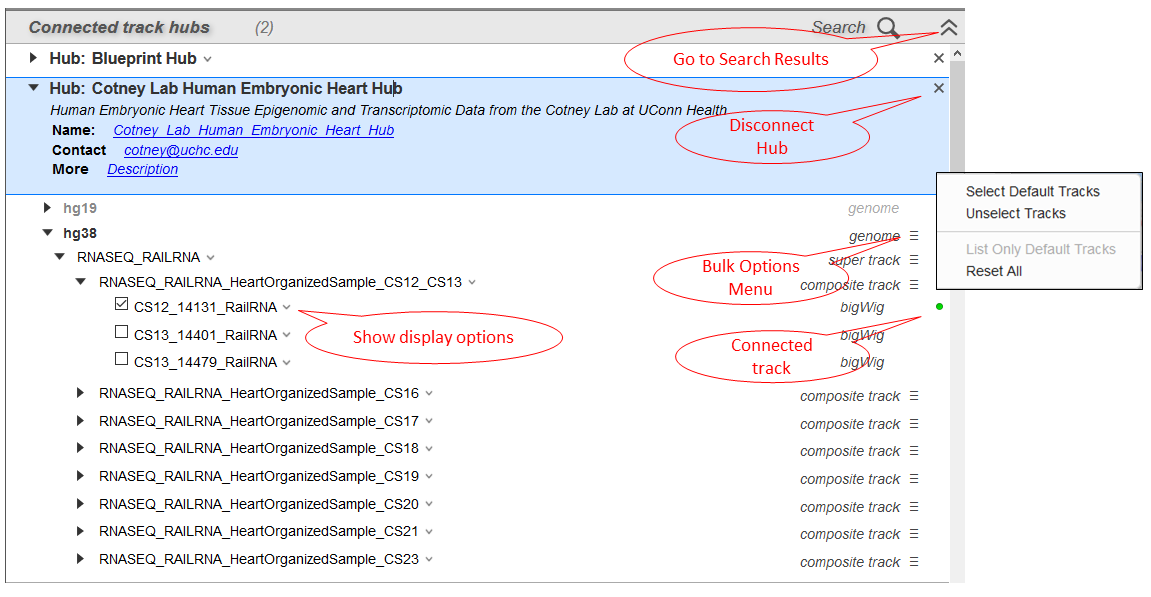
Figure 17. Track Hubs panel configuration view.
Once connected to a track, you can view and adjust display options for that track (Figure 18). Where possible, the track will be pre-configured with default options set by the track hub provider. Display options differ depending on the track data type (e.g. feature, alignment, or graph data).
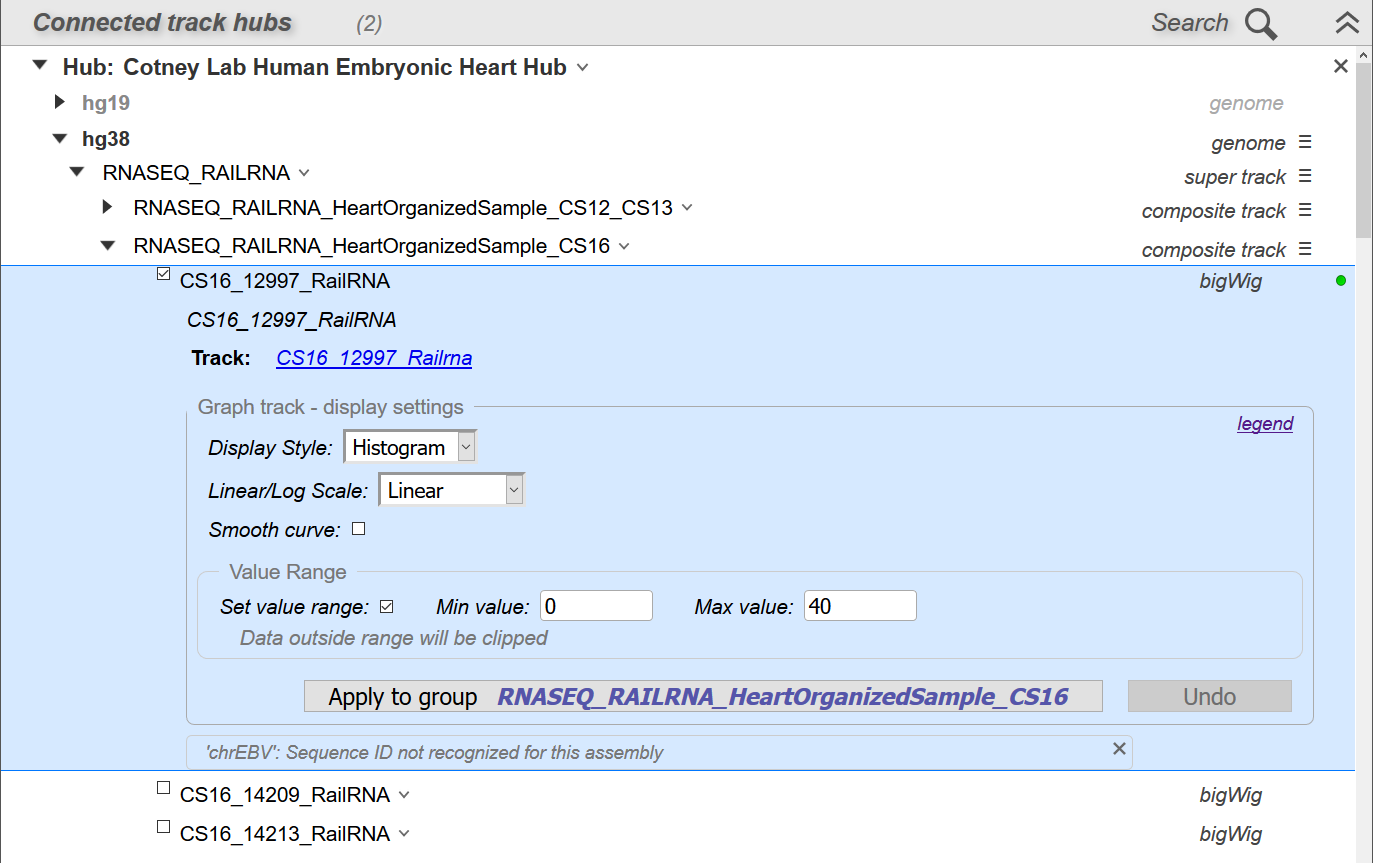
Figure 18. Track Hubs panel configuration view showing display options for a connected track.
Once tracks are connected and configured for display, click on the Configure button in the lower right of the Track Hubs panel to apply changes and view tracks in the Sequence Viewer display.
To disconnect from a track hub, click the x located at the far right of the hub name, and then click the Configure button in the lower right of the dialog. You can also click the Disconnect button in the Tracks and User Data widget. Disconnecting from a hub will also disconnect all tracks from the hub and remove them from the display.
You can also access the Track Hubs panel by selecting the Configure Track Hubs option from the Tracks menu in the Sequence Viewer toolbar.
Track Sets and Collections
The Tracks menu on the Sequence Viewer toolbar also provides access to NCBI Recommended Track Sets (Figure 19). Track Sets are pre-configured assortments of tracks that are commonly viewed together when performing various types of analyses. The names of the track sets indicate the type of analysis for which the track combinations are expected to be most useful. While the content of a Track Set (e.g. Figure 20) is pre-determined, you can still add, remove or re-order the tracks within the display after selecting a Track Set. Any new combinations of NCBI tracks can be saved as a My NCBI Track Collection that you can re-use or share with others.
For more information about Track Sets and Track Collections, please see the following FAQ and the Track Sets and the Track Sets and Track Collections YouTube videos.
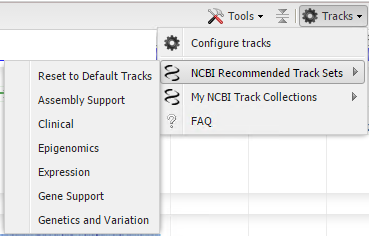
Figure 19. Menu of NCBI Track Sets.
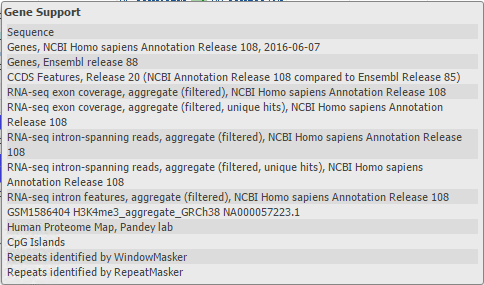
Figure 20. Tracks in the Gene Support track set for the GRCh38 human genome assembly.
BLAST
The BLAST widget (Figure 21A) displays information for BLAST queries (blastn, tblastn) aligned aligned to scaffolds of the assembly displayed in GDV.
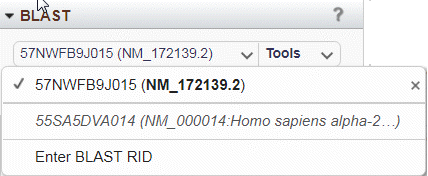
Figure 21A: Select BLAST RID menu. The menu is automatically populated with recent BLAST searches and allows you to enter an active RID.
To load BLAST alignments, open and choose from the Select BLAST RID menu (Figure 21A). You will see the names/RIDs of your recent BLAST searches at NCBI. You can also enter any active BLAST RID. Multiple results can be loaded from this menu; only the results displayed in the widget table will have a check mark next to their name. Click the x next to a BLAST result in this menu to remove it from GDV.
Once loaded to the BLAST widget, the Sequence Viewer display will update to show the corresponding BLAST alignment tracks at top. Track names are provided automatically. Two tracks are provided for each RID: “BLAST results for…” and “Cleaned Alignments-BLAST results for…”. The former provides the raw BLAST hits, while the cleaned alignments track presents a summarized view of co-linear, non-overlapping alignments for each query connected by thin horizontal lines (Figure 21B).
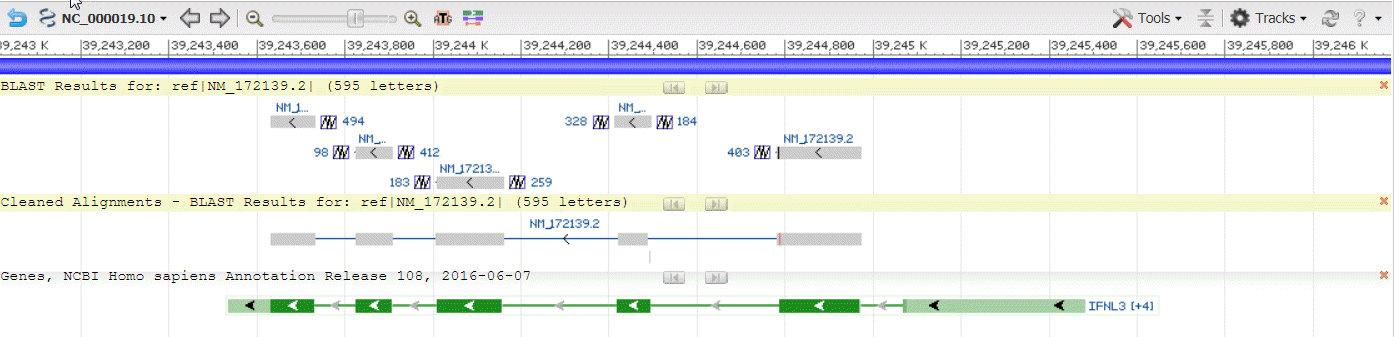
Figure 21B: BLAST alignment tracks. The track name is highlighted in yellow when the corresponding option is selected from the Tools menu.
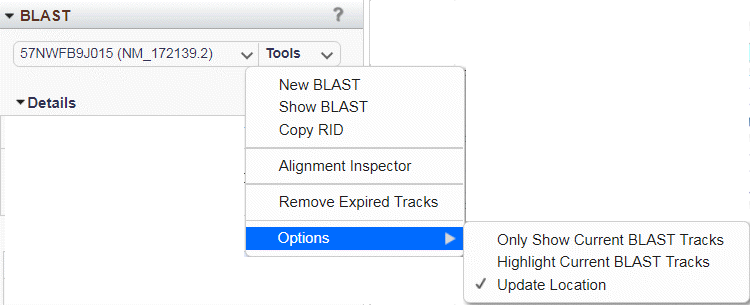
Figure 21C: BLAST Tools menu.
The Tools menu (Figure 21C) displays details and offers additional display options. Use New BLAST to initiate a BLAST search of the viewed genome, or Show BLAST to open a new tab showing the selected BLAST result on the NCBI BLAST website. The Alignment Inspector option will open a more detailed display of the BLAST hits (see below for more information). To remove expired BLAST results from all GDV menus and displays, select Remove Expired Tracks.
The Options sub-menu offers three choices for the display of BLAST alignment tracks. If 'Only Show Current BLAST Tracks' is selected, the Sequence Viewer will only show the alignment tracks corresponding to results loaded to the BLAST widget. Choose the 'Highlight Current BLAST Tracks' option to add a yellow highlight (Figure 21B) to the track titles in the Sequence Viewer. The 'Update Location' option, which is on by default, causes the Sequence Viewer to zoom automatically to the location of a selected BLAST hit.
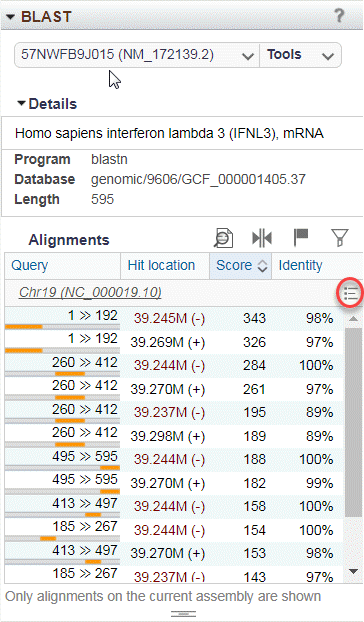
Figure 21D: BLAST Alignments table is populated with BLAST hits from the selected BLAST search.
Expand the Details section to see more information about the query, the BLAST program, and database for the selected BLAST alignment (Figure 21D). The Alignments table summarizes BLAST hits for the selected search. Within the first column of each table row, the query is shown as a gray bar in which the portion shaded orange represents the aligned region. Select a BLAST hit in this table to zoom the Sequence Viewer to the hit location.
BLAST hits can be sorted by any column. The menu on the top right of the table (Figure 21D, red circle) allows you to quickly jump to a location within the table. Hovering over any table row activates a highlight at the corresponding location in the Sequence Viewer. Click one or more rows in the table to select specific BLAST hits. Use the icons above the table or right-click within the table to: (1) zoom in the Sequence Viewer (2) set markers for selections, or (3) filter results by identity.
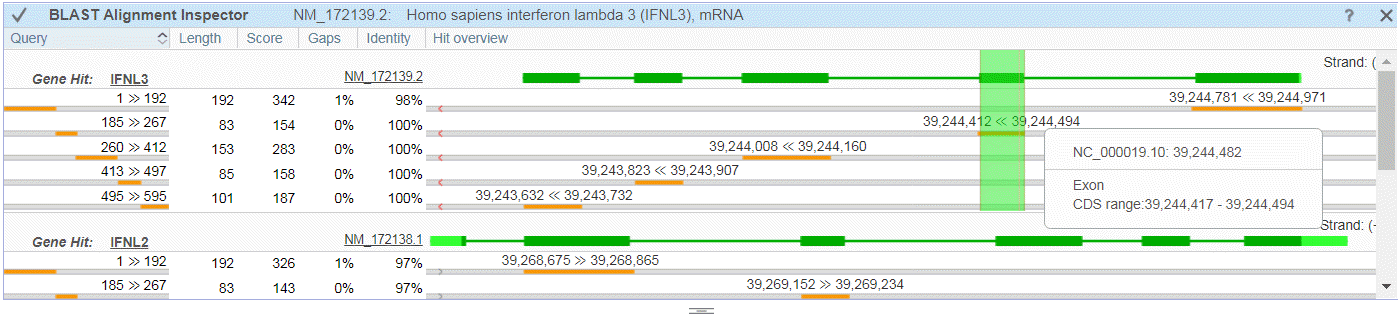
Figure 21E: BLAST Alignment Inspector.
The BLAST Alignment Inspector (Figure 21E) provides a graphical display of the relationship between a BLAST alignment and NCBI's current RefSeq gene annotation. It is launched from within the BLAST widget using the left-most icon above the Alignments table or by right-clicking and selecting the option from the Tools menu.
The Query column in the BLAST Alignment Inspector shows an aligned portion of the query as a shaded orange region. In the Hit Overview column, the genomic coordinates of the aligned region (orange) are shown in relation to a cartoon representing a specific RefSeq transcript. Thick regions of the cartoons indicate exons, thin regions represent introns, and UTRs are shown in light green.
When you place a cursor in the Hit Overview column, you will see a red guide line and tooltip for the corresponding genomic coordinates, including exon-specific details. Hovering over any exon or aligned block highlights this block at the corresponding region of the Sequence Viewer. Click on the block or exon to zoom the Sequence Viewer to that genomic location. Right-click for a menu of additional zoom options or to copy the alignment coordinates.
You can also navigate directly to a Gene or RefSeq transcript in the Sequence Viewer by clicking on the name/accession in the BLAST Alignment Inspector. Note that hovering over icons within the Exon Navigator also creates corresponding highlights in the BLAST Alignment Inspector.
Controlling GDV with URL Parameters
There are several parameters that can be added to the GDV URL. These are typically used when constructing links to GDV from other web pages. Note that GDV requires specification of an id in the URL.
| Parameter | Definition | Example |
|---|---|---|
| id | Identifier (typically accession) from NCBI resource with associated tracks to be displayed in GDV. Required parameter. | Examples:
|
| assm | assembly (RefSeq accession). Note: Do not use if an assembly accession is used for "id" parameter. | https://www.ncbi.nlm.nih.gov/genome/gdv/?id=GSM923418&assm=GCF_000001405.25 |
| context | NCBI resource context that defines default tracks displayed. Examples: GEO, genome, SRA. | https://www.ncbi.nlm.nih.gov/genome/gdv/?id=GSM923418&assm=GCF_000001405.25&context=GEO |
| chr | chromosome number (1-X) or alternate locus | Can specify any chromosome name or top-level accession, e.g. 'chr=1', 'chr=X', 'chr=MT', 'chr=NC_000002.11'. If the "acc" parameter is not supplied, the selected assembly will be applied. Examples:
|
| from/to | range start, end (both 1-based) | Used to specify a genomic range of interest. Requires use of the "chr" parameter. Example:
|
| q | search term (gene, dbSNP id, etc.). Use of this parameter is only encouraged for developers wishing to create links to GDV in which a specific genomic location will be pre-selected for display. | Searches and selects the location of the first search result. Requires explicit specification of assembly in URL. Examples:
|
| mk |
<list-of-marker-spec> - comma separated list of <marker-spec> Creates marker at defined location. Name & color are optional. Region-spec requires an exact match. Use of strings in ranges or positions not supported. |
|
| ts |
ts=<name-pattern> Track Sets The track set name-pattern is case insensitive. It may contain a '*' that will act as a wildcard. Use '%20' to denote a space. User-defined track sets stored in a user's MyNCBI account are supported. |
|
| tsid | tsid is set to an id of a user-defined track set. |
This parameter allows to share a user-defined track set that is made public.
Don't set this parameter directly. Instead, use 'ts' parameter while you are logged in into MyNCBI. GDV will replace the 'ts' parameter with the proper 'tsid=track-set-id'. |