NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Changes in the intracellular Ca2+ concentration affects the visual signal transduction cascade directly or more often indirectly through Ca2+-binding proteins. Here we review recent findings on centrins in photoreceptor cells of the mammalian retina. Centrins are members of a highly conserved subgroup of the EF-hand superfamily of Ca2+-binding proteins commonly associated with centrosome-related structures. In vertebrate photoreceptor cells, centrins are also prominent components in the connecting cilium linking the light-sensitive outer segment with the biosynthetically active inner segment compartment. Recent findings demonstrate that Ca2+-activated centrin forms a complex with the visual G-protein transducin in photoreceptor cells. This Ca2+-dependent assembly of G-proteins with centrin is a novel aspect of the supply of signaling proteins in sensory cells, and a potential link between molecular translocations and signal transduction in general.
Introduction
Vertebrate rod and cone photoreceptor cells are highly specialized, polarized neurons, which consist of morphologically and functionally distinct cellular compartments (see Figs. 1E, 5D). The light-sensitive photoreceptor outer segment is linked with an inner segment via a modified, nonmotile cilium, the so-called connecting cilium. The inner segment contains the organelles typical for the metabolism of an eukaryotic cell and continues into the perikaryon and the synaptic region where the electrical signal generated in the photoreceptor cell is transmitted to secondary neurons of the neuronal retina. The outer segment contains all components of the visual transduction cascade (see below) which are arranged disconnected from the plasma membrane bound to hundreds of stacked membrane discs. These membranous discs are continually renewed throughout lifetime. Newly synthesized membrane is added at the base of the outer segment by the expansion of the plasma membrane1 or by incorporation of vesicular structures into nascent disc membranes,2 whereas discs at the distal tip of the outer segment are phagocyted by the cells of the retinal pigment epithelium.3
At the outer segment disk membrane, photoexcitation of the visual pigment rhodopsin activates a heterotrimeric G protein cascade leading to cGMP hydrolysis in the cytoplasm and closing of cGMP-gated channels (CNG channels) in the plasma membrane.4,5 By its rapid lateral diffusion in the membrane, a single molecule of activated rhodopsin (Rho*) can successively activate hundreds of copies of the tissue-specific G-protein (Gt, transducin, composed of an Gtα subunit bearing the guanine nucleotide binding site with GDP attached and an undissociable βγ-complex), thus amplifying the light signal. The activated, GTP-binding α-subunits holds the effector, a cGMP-specific phosphodiesterase (PDE), in an enzymatically active form before GTP hydrolysis terminates the interaction and the active state of the PDE. As long as active PDE is present, it decreases the cGMP concentration resulting in closure of the CNG channels and a drop of the cationic current through the channels, which is mainly carried by Na and Ca2+. This hyperpolarizes the cell membrane thus providing the neuronal signal by decreasing transmitter release from the synaptic terminal.
The recovery phase of the phototransduction cascade and the adjustment to background light (light adaptation) of photoreceptor cells rely on changes in the intracellular Ca2+concentration, [Ca2+]i. It is well established that changes in [Ca2+]i affects portions of the visual transduction cascade directly or more often indirectly through Ca2+-binding proteins.6 As a consequence of photoabsorption the efflux of Ca2+ (via a light-insensitive plasma membrane Na/CaK exchanger, termed NCKX) exceeds the influx, resulting in [Ca2+]i decrease, which in turn increases the sensitivity of the cGMP-gated channel to cGMP and accelerates the recovery of the dark current by the release of the Ca2+-binding protein calmodulin (CaM) from the β-subunit of the CNG channel (chapter 18 of the present book).7,8 Lowering of [Ca2+]i also stimulates the production of cGMP through activation of a photoreceptor-specific particulate guanylate cyclase (GC).9 The feedback is mediated by one or more Ca2+-binding proteins, termed guanylate cyclase-activating proteins (GCAPs) or GCAP-like proteins (GLPs) described in detail in other chapters of the present book.6 Besides this well-established role of Ca2+ in restoring the dark level of cGMP, yet another mechanism is discussed in the literature, which is thought to act at the level of the activated receptor. It is mediated by another Ca2+-binding protein, recoverin, and affects the phosphorylation of rhodopsin by rhodopsin kinase and thus the quench of light-activated rhodopsin.10 Furthermore, other Ca2+-binding proteins may also regulate the light-insensitive NCKX exchanger.6
The Ca2+-binding proteins involved in the regulation of phototransduction described above are all members of the large EF-hand superfamily of Ca2+-binding proteins which includes besides calmodulin, parvalbumin, troponin C and S100 Ca2+-binding proteins, but also the highly conserved proteins of the centrin subgroup.11,12 We have recently also identified members of the centrin subgroup as structural proteins in vertebrate retinas.13-15 The prominent localization of centrin in cytoskeleton of the connecting cilium of vertebrate photoreceptor cells indicated a role in the intracellular transport between the inner segment and the outer segment of the photoreceptor cell. In view of the importance of Ca2+-binding proteins in the regulation of photoreceptor function, centrin's strategic localization and the small knowledge on centrins in the field will present the most recent information on the centrin subgroup of Ca2+-binding proteins. Besides examining the role of centrins in photoreceptor function, we will also provide new insights, linkages between the signal transduction cascade with the cytoskeleton.
What Are Centrins?
Centrins, also termed “caltractins”, are highly conserved low molecular weight proteins of a subfamily of EF-hand Ca2+-binding proteins.11,12 The first centrin was discovered as the major component of striated flagellar rootlets associated with the basal bodies of unicellular green algae where it participates in Ca2+-dependent and ATP-independent rootlet contractions.16 Centrins have since been found to be ubiquitously associated with centrioles of basal bodies and centrosomes, and mitotic spindle poles in cells from diverse organisms, including yeast, ciliates, green algae, higher plants, invertebrates, and vertebrates (Fig. 1).11,12
Centrin Genes and Molecular Structure of Centrin Proteins
Cloning efforts in recent years have resulted in the identification of centrin genes in a variety of species from all kingdoms of eukaryotic organisms, protists, fungi, plants, and animals.17-27 Analyses of amino acid sequences deduced from the cDNA clones demonstrate that centrins are a highly conserved, yet distinct subfamily of the EF-hand superfamily of Ca2+-binding proteins (Fig. 2). Centrins are acid proteins, about 170 amino acids in length, which is in good agreement with their apparent molecular mass of about 20 kDa.11,12 To date, in lower eukaryotes like the yeast Saccharomyces cerevisiae or the unicellular green algae Chlamydomonas reinhardtii only one centrin gene (ScCDC31 and CrCEN, respectively) has been identified, whereas in the genome of vertebrates at least three centrin genes (e.g., HsCEN1, HsCEN2, and HsCEN3) are present.17-19,21,22,26 Clustal analyses of deduced amino acid sequences of centrins from different organisms reveal several phylogenetic groups of centrins (Fig. 3). While some protist centrin species can not be grouped into homogeneous groups, most centrins of higher plants, green algae centrins, and all three known vertebrate centrin isoforms form a phylogenetic group. In vertebrates, Cen1p isofoms and Cen2p isoforms are very closely related showing amino acid identities of about 80 percent to 90 percent, whereas sequences of the yeast centrin (ScCdc31p)-related vertebrate Cen3p isoforms have only amino acid identities of about 55 percent to both other isoforms. Interestingly, in vertebrate species Cen1p and Cen2p isoforms are closer related to algal centrin (e.g., CrCenp) than to Cen3p isoform of the same species, strongly suggesting two divergent subfamilies.26
As members of the parvalbumin superfamily of Ca2+-binding proteins, centrins contain four helix-loop-helix EF-hands consensus domains which may each bind one Ca2+.28-31 Protein sequence comparisons between different centrin species reveal that the EF-hand consensus motifs are the most highly conserved domains (Fig. 2). Further phylogenetic analyses indicate that the EF-hand domains arose from twofold duplication of an ancestral EF-hand motif.32 However, during molecular phylogenesis EF-hand motifs in centrins lost their ability to bind Ca2+. Binding studies indicate that in the green algae Chlamydomonas CrCenp and Tetraselmis TsCenp all four EF-hands bind a Ca2+, two EF-hands bind Ca2+with high affinity and two EF-hands bind Ca2+with low affinity,33,34 whereas other green algae possess two or three functional EF-hands.27 Sequence analysis of vertebrate centrin isoforms suggests that Cen1p and Cen2p molecules bind two Ca2+with their first and the fourth EF-hand and in Cen3p the fourth is the last remaining functional EF-hand motif as it is the case in the yeast centrin ScCdc31p.11,26,27,35 There are several lines of evidence that Ca2+-binding to centrins induces drastic conformation changes in centrin molecules 11,12,36,37 as previously demonstrated for the related EF-hand protein calmodulin.38-40 In contrast to calmodulin, centrin molecules become more compact upon Ca2+-binding and Ca2+-activated centrins form dimers and oligomers.36,37 In polymerization assays, Ca2+-binding induces even centrin polymers, not only with green algae centrins but also with mammalian centrin 1,36 which may be the structural basis for contractile centrin-fiber systems (see above). Furthermore, Ca2+-binding to centrins increases the affinity of centrin-binding proteins to centrins 36,37,41 which we recently also demonstrated in mammalian photoreceptor cells (Gießl et al, in preparation).35 To understand Ca2+-induced conformation changes in centrins and binding characteristics of target proteins of centrins, data from high resolution structural analysis are required.
The amino-terminal subdomain of centrins is unique for small Ca2+-binding proteins, unlike those found in, e.g., calmodulin or GCAPs. It is also the most distinctive and variable region of centrins and it has been suggested to be responsible for some functional diversity among centrin species.11,32,36 Studies on the polymerization properties of centrins indicate that the Ca2+-induced polymerization of centrins, e.g., the formation of contractile centrin-fibers in green algae, is mainly dependent on the amino-terminal domain.36 In the green algae, it has been demonstrated that centrin phosphorylation correlates with centrin-fiber elongation (relaxation).16,42 Although conserved potential sites for phosphorylation by protein kinase A (PKA) and p34cdc2 kinase are located in the amino-terminal of centrins,11 direct evidence for in vivo phosphorylation at the amino-terminus of centrins is missing. However, aberrant centrin phosphorylation has been shown under pathogenic conditions in human breast cancer cells that have amplified centrosomes containing supernumerary centrioles.43 Furthermore, recent studies by Lutz and coworkers44 indicate that vertebrate centrins are phosphorylated by PKA at conserved PKA consensus sequences present in the carboxy-terminal of centrin molecules. These results suggest that centrin phosphorylation in centrioles signals the separation of centrosomes during the prophase of the cell cycle.
Centrin's Cellular Localization and Function
Centrin was first described as the major component of the massive striated flagellar rootlets of the Prasinophocean unicellular green alga Tetraselmis striata.16 Centrin-containing striated rootlets are commonly found in unicellular green algae.45 They originate near the centrioles of the the basal body apparatus, project into the cytoplasm of the cell body and extend to the plasma membrane, the nucleus or other organelles. In the algal model system Chlamydomonas, descending centrin-based fibers connect the basal body apparatus with the nucleus (Fig. 1A).46,47 In addition to these descending fibers, in Chlamydomonas at least two other fiber systems contain centrin: the distal fibers which connect both adjacent basal bodies to one another48 and the stellate fibers in the transition zone in the plane between the basal body and the axoneme of the flagella.49 In green algae, all of these centrin fibers have in common that they contract in response on an increase of the intracellular Ca2+ concentration, [Ca2+]i. Most interestingly, Ca2+-triggered contraction of centrin fibers of the transition zone may induce microtubule severing and thereby the excision of the flagellum.49,50 Present microtubule severing mediated by Ca2+-activated centrin may be a more widespread phenomenon preceding the massive reorganization of the microtubule cytoskeleton during cell migration51 or contributing to the microtubule released from the centrosome, the major microtubule organizing center (MTOC) of higher eukaryotic cells.52
Major contributions to evaluate the function of centrins in the cell cycle are provided by intensive studies on yeast centrin.12,36,41,53,54 In baker's yeast S. cerevesiae, centrin encoded by the CDC31 gene functions in the duplication of the spindle pole body, the structural equivalent of the centrosome in higher eukaryotic cells. During the first steps of the yeast spindle pole body duplication the binding of Cdc31p to Kar1p is required. Furthermore, Cdc31p specifically interacts with other yeast proteins including an essential kinase (Kic1p) which activity probably regulates the spindle pole body duplication.53,55
In vertebrates, centrin proteins are ubiquitously expressed commonly associated with centrosome-related structures such as spindle poles of dividing cells or centrioles in centrosomes and basal bodies.11,12 As discussed above, in mammals at least three centrin genes are expressed which may cluster to two divergent subfamilies.26 As a consequence of the isoform diversity the three mammalian centrin isoforms may also exhibit differences in their subcellular localization as well as in their cellular function. Unfortunately, little is known about the specific subcellular localization of the different centrin isoforms in diverse cell types and tissues. Most studies on the localization of the centrin in mammalian cells and tissues have been performed with polyclonal and monoclonal antibodies raised against green algae centrins which do not discriminate between the centrin isforms. Using these antibodies, centrins were detected in the centrioles of centrosomes and in the pericentriolar matrix.56-58 Further immunological experiments show that antibodies to yeast Ccd31p or mammalian Cen3p react exclusively with Cen3p26,59 whereas, to our knowledge, to date all of the antibodies raised against the closely related mammalian Cen1p or Cen2p isoforms react with both isoforms59 (Gießl et al in preparation). Nevertheless, recent studies by immunoelectron microscopy demonstrate that Cen1p/Cen2p and Cen3p are localized in the central lumen of the centrioles of centrosomes and basal bodies.59,60 In human ciliated tracheal cells, immunoelectron microscopy reveals that the isoform Cen3p is exclusively a core component of the basal body centriole, antibodies to Cen1p/Cen2p additionally decorate epitopes in transition zone of motile cilia.59 Furthermore, comparative RT-PCR experiments (combined reverse transcriptase reaction and polymerase chain reaction) using isoform-specific primers demonstrate that CEN2 is ubiquitously expressed, whereas CEN1 expression is restricted to ciliated cells.15,59 Thus, it is likely that Cen1p functions as a centrin isoform in compartments of cilia and flagella. Functional analyses indicate that ciliary centrin are involved in the beating of cilia which is controlled by the intraciliary Ca2+ concentration.59
The prominent localization of centrins at the centrosomes and basal bodies gave the rise for several hypothesis of the function of centrins. In interphase cells or in arrested cells of differentiated tissue, the centrosome functions as the major microtublule organizing center determining the number and polarity of cytoplasmic microtubules. Polymerization of novel microtubules at the centrosome is preceded by the de novo nucleation of microtubules in the pericentriolar matrix that surrounds and connects the centriole pair of an individual centrosome. It has been suggested that centrins are involved in the microtubule severing which should occur to release de novo synthesized microtubules from the pericentriolar origin.52 However, more reliable evidence was gathered that centrins may play important, but probably distinct roles at the centrosome during the cell cycle. Once in the cell cycle, the centrosome is duplicated to give rise to two spindle poles that organize the microtubules array of the mitotic spindle. While Cen3p, as its yeast relative Cdc31p, participates in centrosome reproduction and duplication,61 Cen1p/Cen2p may play a role in centriole separation preceding centrosome duplication.44
Centrins in the Vertebrate Retina
RT-PCR studies with centrin isoform-specific primers reveal that all three centrin isoforms are expressed in the mammalian retina, which has been confirmed by Western blot analysis using antibodies specific for Cen3p and Cen1p/Cen2p, respectively15 (A. Gießl, A. Schmitt, and U. Wolfrum, unpublished results). Further studies showed that centrins are expressed in the retina of species distributed throughout the subphylum of vertebrates (Fig. 4). Thus, centrins are probably ancient cytoskeletal proteins in the vertebrate retina indicating this conserved basic function in retinal cells.
Immunocytochemical studies demonstrate that centrins are concentrated in the cells of the vertebrate retina in two basically distinct structural domains (Fig. 5). As in other cell types of animals, centrins are components of the centrioles of centrosomes and basal bodies in the retinal neurons contributing to centrosome function discussed above. However, in all of our studies on numerous different vertebrate species, indirect anti-centrin immunofluorescene was most prominent in photoreceptor cell layer (Fig. 5).13-15
Centrin Functions as a Cytoskeletal Component of the Connecting Cilium of the Photoreceptor Cell
Higher magnification of anti-Cen1p/Cen2p stained cryosections through vertebrate retinas shows that centrins are localized at the photoreceptor layer at the joint between the photoreceptor inner segment and outer segment (Fig. 5). Analysis of immunolabled isolated photoreceptor fragments reveals that centrins are not only present in the basal body, but also localized along the entire longitudinal extension of the connecting cilium (Fig. 5E).14,15 Precise subcellular localization by immunoelectron microscopy and the quantification of silver-enhanced immunogold labeling show that centrin is localized in the subciliary domain of the inner face of the microtubule doublets of the connecting cilium of rod and cone photoreceptor cells (Fig. 6).35 As in other ciliated cells, in photoreceptor cells the centrin decorated by immunolabeling in the connecting cilium most likely resembles the centrin 1 isoform.
The modified connecting cilium of vertebrate photoreceptor cells is the structurally equivalent of an extended transition zone present at the base of a common motile cilium.62 Therefore, the presence of centrin (most probably centrin 1) along the entire extension of the connecting cilium of photoreceptor cells is in agreement with the centrin localization in the transition zone of motile cilia or the sensory cilia of mammalian olfactory cells.59 In photoreceptor cells, the connecting cilium links the morphological and functional distinct cellular compartments of the light-sensitive outer segment with the biosynthetically active inner segment. The connecting cilium serves as an active barrier for membrane components and soluble proteins regulating free diffusion between the inner and the outer segment of photoreceptor cells.62,63 Since it is also the only intracellular bridge between both segments, intracellular exchanges between the inner segment and the outer segment are forced to occur through the slender connecting cilium.62 Recently, we and others have shown that the visual pigment opsin is translocated to its final destination at the base of the photoreceptor outer segment along the membrane of the connecting cilium.64-66 Actin filament-based and microtubule-associated transport processes seem to be involved in the unidirectional ciliary transport of opsin: The membrane associated molecular motor protein myosin VIIa has been shown to participate in ciliary transport of rhodopsin.64-66 Marszalek and co-workers67 gathered indications by a genetic approach that the microtubule-based heterotrimeric kinesin II-motor might be additionally involved in ciliary transport of rhodopsin but also of arrestin. However, cytoskeletal molecules associated with other proteins of the visual transduction cascade and which, therefore, are probably involved in the ciliary translocation of these proteins, have not yet been identified. The prominent localization of centrin in the connecting cilium of photoreceptor cells obviously indicates a specific role of centrin in the function of the photoreceptor cilium. Besides its possible role in ciliary transport, an involvement of centrin in retinomotor movement and in the photoreceptor outer segment alignment or orientation has been discussed.14 If any of these processes are based on the centrin system of the cilium they should be dependent on and regulated by changes of the intracellular Ca2+ concentration. Our recent results, as discussed below provide striking evidence for Ca2+-dependent interaction between centrin 1 and the visual G-protein transducin on its pathway through the inner lumen of connecting cilium of mammalian photoreceptor cells.35
Centrin-Interacting Proteins in Mammalian Photoreceptor Cells
In the context of the cell, protein function and its regulation is determined by the binding proteins to the target protein. Unfortunately, little is known about centrin-binding proteins in mammalian photoreceptor cells. To evaluate centrin functions in vertebrates, in other experimental systems, different strategies for the identification of centrin-associated or centrin-interacting proteins were applied. Analysis of proteins in coimmunoprecipitations performed with antibodies against algae centrin has revealed centrin interaction with the heat shock proteins HSP70 and HSP90 in cytoplasm of arrested Xenopus oocytes.68 The centrin/HSP complex may sequestrate centrin in a nonactive form until Ca2+ activation of the oocyte causes the dissociation of the complex making centrin available for subsequent centrosome assembly. In yeast 2-hybrid screens the laminin-binding protein (LBP) of the basal lamina and the cytoplasmic receptor protein tyrosine kinase k have been identified as proteins interacting with HsCen2p, the ubiquitously expressed centrin isoform.69 Although, there is no specific experimental evidence, none of the proteins identified as centrin-binding proteins has an obvious function in the connecting cilium of the photoreceptor cells. Nevertheless, Western blot overlay assays of retinal proteins with recombinant expressed MmCen1p indicate the presence of several centrin 1-binding proteins in the mammalian retina (Fig. 7). However, only centrin 1 in its Ca2+-activated form interacts with the several polypeptides. A Ca2+-dependent increase of the affinity of centrin to target proteins is known from binding of the yeast centrin Cdc31p to Kar1p12 which has been confirmed in in vitro binding studies of diverse recombinant expressed centrin species to the yeast target protein.36,41 Further analysis of the proteins identified by the MmCen1p overlay assay are currently performed. However, the centrin 1-binding protein p37 has been already identified as the β-subunit of the visual G-protein transducin (Gt) (Fig. 8,9B).
Centrin/Transducin Complex
Recently, evidence was provided that MmCen1p interacts with the visual G-protein transducin with high affinity, and thereby form functional protein-protein complexes in photoreceptor cells in a Ca2+-dependent manner.35 Transducin is the tissue-specific G-protein of the visual signal transduction cascade of the photoreceptor cells in the vertebrate retina (see also introduction). Upon light-activation, rhodopsin (Rho*) activates hundreds of G-protein molecules and the light signal is amplified. This receptor-G-protein interaction requires the intact Gt holoprotein, composed of an α-subunit bearing the guanine nucleotide binding site with GDP bound and an undissociable βγ-complex, and initiates the intermolecular transduction of the light signal by catalyzing the exchange of GDP for GTP in the α-subunit of the G-protein. Activated, GTP-binding α-subunits are free to couple to the effector, a cGMP-specific phosphodiesterase (PDE).
In vertebrate photoreceptor cells the subcellular localization of transducin is modulated by light: in the dark Gt is highly concentrated in outer segments while in light, the majority of Gt is translocated and abundantly localized in the inner segment and the cell body of photoreceptor cells (Fig. 8).35,70,71 Light-induced exchanges and movements of the cytoplasmic components between the photoreceptor segments have to occur through the connecting cilium, since the slender cilium serves as the only intracellular linkage between both photoreceptor compartments. As described above, centrin 1 is a prominent component of the cytoskeleton of the nonmotile modified cilium and immunufluorscence double labelings of tranducin and centrin indicate that transducin and centrin 1 co-localize in the connecting cilium (Fig. 8C). Further immunoelectron microscopical analysis and the quantification of silver-enhanced immunogold decorations reveal that centrin and transducin are not only exist parallel in the cilium, but share also the same subciliary domain, the inner ciliary lumen of the connecting cilium.35 Their spatial co-distribution indicate that both proteins may physically interact during the exchange of transducin between the photoreceptor segments through the cilium.
Recently, we have gathered striking evidence that centrin 1 and transducin indeed interact with high affinity.35 In vitro assays including co-immunoprecipitation, overlay and cosedimentation assays as well as size-exclusion chromatography and kinetic light-scattering experiments independently demonstrate that centrin 1 binds with high affinity to transducin (Fig. 9).35 Our studies also show that the protein-protein interaction centrin 1 and transducin is highly specific: centrin 1 specifically interacts with transducin and does not bind to other components of the visual signal transduction cascade (e.g., arrestin, rhodopsin, rhodopsin kinase, PDE). The centrin relatives recoverin and calmodulin do not show significant affinities to transducin. The analysis of MmCen1p overlay assays with antibodies specific to transducin subunites and size-exclusion chromatographies further demonstrate that assembly of centrin 1/G-protein complex is mediated by the βγ-complex (see also Fig. 9B, DG). Our data also reveal that the assembly of the centrin 1/G-protein protein complex is strictly dependent on the Ca2+ concentration and that at least two Ca2+ ions are required for the activation of centrin 1 necessary for the formation of centrin 1/G-protein complex. Moreover, further analysis indicates that activated centrin 1 binds as a homooligomer to the βγ-complex of transducin.35
What is the role of the centrin 1/G-protein complex in the photorecptor cell? Current working hypotheses of the centrin 1/G-protein complex function in the photorecptor cell are summarized in the cartoon in figure 10. The spatial colocalization of centrin 1 and transducin in the lumen of the connecting cilium emphasizes that in photoreceptor cells, the formation of centrin 1/G-protein complex should occur in this ciliary compartment. An increase of the intracellular Ca2+concentration in the photoreceptor cell should cause the activation of centrin 1 in the connecting cilium and in turn induce the binding of centrin 1 oligomers to transducin passing through the ciliary. As a consequence of the assembly of centrin/transducin complexes the movement of transducin should be effected. In photoreceptors, light-modulated changes of free Ca2+ in the outer segment which include the well-studied Ca2+ drop within the operating (single quantum detective) range of the rod10 and recent observations of Ca2+ increase in bright light (rod-saturated conditions)72 should also effect the free Ca2+ in the connecting cilium. In the cilium the assembly of centrin 1/G-protein complexes may contribute to a Ca2+induced barrier for further exchange of transducin between the photoreceptor inner and outer segment (barrier hypothesis Fig. 10B). A drop of Ca2+ should induce the disassembly of the complex, thus providing a necessary condition for the light-modulated exchange of transducin between the inner and the outer segment of photoreceptor cells described above.35,70,71 However, Ca2+-triggered sequential binding of transducin to centrin 1 may also contribute to the transport of transducin though the photoreceptor connecting cilium (Ca2+ gradient hypothesis Fig. 10C). The Ca2+-dependent assembly of a G-protein with centrin is a novel aspect of the supply of signaling proteins in sensory cells, and a potential link between molecular translocations and signal transduction in general.
Conclusion
Centrins are members of a conserved subfamily of EF-hand Ca2+-binding proteins. During the past years, 3 centrin isoforms have been found to be ubiquitously associated with the centrioles of centrosomes or centrosome-related structures in diverse vertebrate cells. Our work on centrins in photoreceptor cells has revealed that centrins are prominent components of the ciliary apparatus of photoreceptor cells. Although several lines of evidence indicate defined spatial distributions of the known centrin isoforms, the differential localization of centrin isoforms by using isoform-specific antibodies or by the transfection of the retina with tagged-centrin constructs should provide more reliable information on the specific localization and function of the centrin isoforms in photoreceptor cells. Our recent findings reveal that the centrin isoform centrin 1 binds with high affinity to transducin in a strict Ca2+-dependent manner. Additional experimental efforts are necessary to resolve the question whether transducin binding is restricted to centrin 1. If so, what are the functions of the other centrin isoforms in photoreceptor cells? The results of the current analysis of putative centrin-associated proteins (other than transducin) in the mammalian retina will most probably also provide further insights in the role of centrins in photoreceptor cell function. In the future, we will also address the question whether Ca2+ activation of centrins is the only post-translational modification regulating the function(s) of centrin in photoreceptor cells. And finally, the clarification of the structure of centrin isoforms will also enlighten the molecular mechanisms of the diverse functions of centrins in photoreceptors.
Acknowledgements
This work was supported by grants of the Deutsche Forschungsgemeinschaft (DFG) to U.W. (Wo548/1), the DFGSPP 1025 “Molecular Sensory Physiology” to A.P. (Ho832/6) and U.W. (Wo548/3), and the FAUN-Stiftung, Nürnberg, Germany to U.W. The authors thank Dr. K. P. Hofmann, HumboldtUniversität, Universitätsklinikum Charité, Berlin, Germany, for helpful comments and critical discussions of the manuscript and Brenda K. Huntley, Mayo Clinic Foundation, Rochester, MN, USA, for attentive linguistic corrections. We also thank Dr. H. E. Hamm, Northwestern University Institute of Neuroscience, University of Texas at Austin, USA, for kindly supplying the monoclonal antibody to frog α-transducin and to Dr. J. L. Salisbury, Mayo Clinic Foundation, Rochester, MN, USA, for providing us mouse centrin 1 cDNA and antibodies raised against algae centrin.
References
- 1.
- Steinberg RH, Fisher SK, Anderson DH. Disc morphogenesis in vertebrate photoreceptors. J Comp Neurol. 1980;190:501–508. [PubMed: 6771304]
- 2.
- Usukura J, Obata S. Morphogenesis of photoreceptor outer segments in retinal development. Prog Retin Eye Res. 1995;15:113–25.
- 3.
- Young RW. Visual cells and the concept of renewal. Invest Ophthalmol. 1976;15:700–25. [PubMed: 986765]
- 4.
- Heck M, Hofmann KP. G-protein-effector coupling: a real-time light-scattering assay for transducin-phosphodiesterase interaction. Biochemistry. 1993;32:8220–7. [PubMed: 8394130]
- 5.
- Okada T, Ernst OP, Palczewski K. et al. Activation of rhodopsin: new insights from structural and biochemical studies. Trends Biochem Sci. 2001;26:318–24. [PubMed: 11343925]
- 6.
- Palczewski K, Polans AS, Baehr W. et al. Ca2+-binding proteins in the retina: structure, function, and the etiology of human visual diseases. Bioessays. 2000;22:337–50. [PubMed: 10723031]
- 7.
- Hsu YT, Molday RS. Modulation of the cGMP-gated channel of rod photoreceptor cells by calmodulin. Nature. 1993;361:76–9. [PubMed: 7678445]
- 8.
- Weitz D, Zoche M, Muller F. et al. Calmodulin controls the rod photoreceptor CNG channel through an unconventional binding site in the N-terminus of the beta-subunit. EMBO J. 1998;17:2273–84. [PMC free article: PMC1170571] [PubMed: 9545240]
- 9.
- Koch KW, Stryer L. Highly cooperative feedback control of retinal rod guanylate cyclase by calcium ions. Nature. 1988;334:64–6. [PubMed: 2455233]
- 10.
- Molday RS, Kaupp UB. Ion channels of vertebrate photoreceptorsIn: Stavenga DG, Degrip WJ, Pugh ENJr, eds. Molecular Mechanism in Visual Transduction Amsterdam: Elsevier Science Publishers,2000143–182.
- 11.
- Salisbury JL. Centrin, centrosomes, and mitotic spindle poles. Curr Opinion Cell Biol. 1995;7:39–45. [PubMed: 7755988]
- 12.
- Schiebel E, Bornens M. In search of a function for centrins. Trends Cell Biol. 1995;5:197–201. [PubMed: 14731449]
- 13.
- Wolfrum U. Cytoskeletal elements in arthropod sensilla and mammalian photoreceptors. Biol Cell. 1992;76:373–381. [PubMed: 1305480]
- 14.
- Wolfrum U. Centrin in the photoreceptor cells of mammalian retinae. Cell Motil Cytoskeleton. 1995;32:55–64. [PubMed: 8674134]
- 15.
- Wolfrum U, Salisbury JL. Expression of centrin isoforms in the mammalian retina. Exp Cell Res. 1998;242:10–17. [PubMed: 9665797]
- 16.
- Salisbury JL, Baron A, Surek B. et al. Striated flagellar roots: isolation and characterization of a calcium-modulated contractile organelle. J Cell Biol. 1984;99:962–70. [PMC free article: PMC2113404] [PubMed: 6381510]
- 17.
- Huang B, Mengerson A, Lee VD. Molecular cloning of cDNA for caltractin, a basal body-associated Ca2+-binding protein: homology in its protein sequence with calmodulin and the yeast CDC31 gene product. J Cell Biol. 1988;107:133–40. [PMC free article: PMC2115161] [PubMed: 2839516]
- 18.
- Baum P, Furlong C, Byers BE. Yeast gene required for spindle pole body duplication: homology of its product with Ca2+-binding proteins. Proc Natl Acad Sci USA. 1986;83:5512–5516. [PMC free article: PMC386317] [PubMed: 3526331]
- 19.
- Baum P, Yip C, Goetsch L. et al. A yeast gene essential for regulation of spindle pole duplication. Mol Cell Biol. 1988;8:5386–97. [PMC free article: PMC365641] [PubMed: 3072479]
- 20.
- Zhu JA, Bloom SE, Lazarides E. et al. Identification of a novel Ca2+-regulated protein that is associated with the marginal band and centrosomes of chicken erythrocytes. J Cell Sci. 1995;108:685–98. [PubMed: 7769011]
- 21.
- Lee VD, Huang B. Molecular cloning and centrosomal localization of human caltractin. Proc Natl Acad Sci USA. 1993;90:11039–43. [PMC free article: PMC47917] [PubMed: 8248209]
- 22.
- Errabolu R, Sanders MA, Salisbury JL. Cloning of a cDNA encoding human centrin, an EF-hand protein of centrosomes and mitotic spindle poles. J Cell Sci. 1994;107:9–16. [PubMed: 8175926]
- 23.
- Meng TC, Aley SB, Svard SG. et al. Immunolocalization and sequence of caltractin/centrin from the early branching eukaryote Giardia lamblia. Mol Biochem Parasitol. 1996;79:103–8. [PubMed: 8844677]
- 24.
- Madeddu L, Klotz C, Le Caer JP. et al. Characterization of centrin genes in Paramecium. Eur J Biochem. 1996;238:12–18. [PubMed: 8665928]
- 25.
- Levy YY, Lai EY, Remillard SP. et al. Centrin is a conserved protein that forms diverse associations with centrioles and MTOCs in Naegleria and other organisms. Cell Motil Cytoskeleton. 1996;33:298–323. [PubMed: 8801035]
- 26.
- Middendorp S, Paoletti A, Schiebel E. et al. Identification of a new mammalian centrin gene, more closely related to Saccharomyces cerevisiae CDC31 gene. Proc Natl Acad Sci USA. 1997;94:9141–9146. [PMC free article: PMC23077] [PubMed: 9256449]
- 27.
- Wottrich R. Klonierung und computergestützte Strukturanalyse von Centrinisoformen der Ratte (Rattus norvegicus). Cloning and computer based structural analysis of centrin isoforms in the rat (Rattus norvegicus ) Diploma thesis 1998. University of Karlsruhe, Germany.
- 28.
- Kretsinger RH. Evolution and function of calcium-binding proteins. Int Rev Cytol. 1976;46:323–93. [PubMed: 186427]
- 29.
- Kretsinger RH. Calcium-binding proteins. Annu Rev Biochem. 1976;45:239–66. [PubMed: 134666]
- 30.
- Moncrief ND, Kretsinger R, Goldman M. Evolution of EF-hand calcium-modulated proteins. I. Relationships based on amino acid sequences. J Mol Evol. 1990;30:522–62. [PubMed: 2115931]
- 31.
- Nakayama S, Moncrief ND, Kretsinger RH. Evolution of EF-hand calcium-modulated proteins. II. Domains of several subfamilies have diverse evolutionary histories. J Mol Evol. 1992;34(5):416–48. [PubMed: 1602495]
- 32.
- Bhattacharya D, Steinkötter J, Melkonian M. Molecular cloning and evolutionary analysis of the calcium-modulated contractile protein, centrin, in green algae and land plants. Plant Mol Biol. 1993;23(6):1243–54. [PubMed: 8292788]
- 33.
- Coling DE, Salisbury JL. Characterization of the calcium-binding contractile protein centrin from Tetraselmis striata (Pleurastrophyceae). J Protozool. 1992;39:385–91. [PubMed: 1640386]
- 34.
- Weber C, Lee VD, Chazin WJ. et al. High level expression in Escherichia coli and characterization of the EF-hand calcium-binding protein caltractin. J Biol Chem. 1994;269:15795–802. [PubMed: 8195234]
- 35.
- Pulvermüller A, Gießl A, Heck M. et al. Calcium-dependent assembly of centrin-G-protein complex in photoreceptor cells. Mol Cell Biol. 2002;22:2194–2203. [PMC free article: PMC133667] [PubMed: 11884606]
- 36.
- Wiech H, Geier BM, Paschke T. et al. Characterization of green alga, yeast, and human centrins. Specific subdomain features determine functional diversity. J Biol Chem. 1996;271:22453–61. [PubMed: 8798410]
- 37.
- Durussel I, Blouquit Y, Middendorp S. et al. Cation- and peptide-binding properties of human centrin 2. FEBS Lett. 2000;472:208–12. [PubMed: 10788612]
- 38.
- Barbato G, Ikura M, Kay LE. et al. Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: the central helix is flexible. Biochemistry. 1992;31(23):5269–78. [PubMed: 1606151]
- 39.
- Meador WE, Means AR, Quiocho FA. Modulation of calmodulin plasticity in molecular recognition on the basis of x-ray structures. Science. 1993;262(5140):1718–21. [PubMed: 8259515]
- 40.
- Crivici A, Ikura M. Molecular and structural basis of target recognition by calmodulin. Annu Rev Biophys Biomol Struct. 1995;24:85–116. [PubMed: 7663132]
- 41.
- Geier BM, Wiech H, Schiebel E. Binding of centrins and yeast calmodulin to synthetic peptides corresponding to binding sites in the spindle pole body components Kar1p and Spc110p. J Biol Chem. 1996;271:28366–74. [PubMed: 8910460]
- 42.
- Martindale VE, Salisbury JL. Phosphorylation of algal centrin is rapidly responsive to changes in the external milieu. J Cell Sci. 1990;96:395–402. [PubMed: 2229193]
- 43.
- Lingle WL, Lutz WH, Ingle JN. et al. Centrosome hypertrophy in human breast tumors: implications for genomic stability and cell polarity. Proc Natl Acad Sci USA. 1998;95(6):2950–5. [PMC free article: PMC19675] [PubMed: 9501196]
- 44.
- Lutz W, Lingle WL, McCormick D. et al. Phosphorylation of centrin during the cell cycle and its role in centriole separation preceding centrosome duplication. J Biol Chem. 2001;276(23):20774–80. [PubMed: 11279195]
- 45.
- Salisbury JL. Centrin and the algal flagellar apparatus. J Phycol. 1989;25:201–6.
- 46.
- Salisbury JL, Sanders MA, Harpst L. Flagellar root contraction and nuclear movement during flagellar regeneration in Chlamydomonas reinhardtii. J Cell Biol. 1987;105:1799–805. [PMC free article: PMC2114663] [PubMed: 3667698]
- 47.
- Schulze D, Robenek H, McFadden GI. et al. Immunolocalization of a Ca2+modulated contractile protein in the flagellar apparatus of green algae: the nucleusbasal body connector. Eur J Cell Biol. 1987;45:51–61.
- 48.
- McFadden GI, Schulze D, Surek B. et al. Basal body reorientation mediated by Ca2+-modulated contractile protein. J Cell Biol. 1987;105:903–12. [PMC free article: PMC2114749] [PubMed: 3305524]
- 49.
- Sanders MA, Salisbury JL. Centrin-mediated microtubule serving during flagellar excision in Chlamydomonas reinhardtii. J Cell Biol. 1989;108:1751–60. [PMC free article: PMC2115546] [PubMed: 2654141]
- 50.
- Sanders MA, Salisbury JL. Centrin plays an essential role in microtubule severing during flagellar excision in Chlamydomonas reinhardtii. J Cell Biol. 1994;124:795–805. [PMC free article: PMC2119956] [PubMed: 8120100]
- 51.
- Salisbury JL. Algal centrin: Calcium sensitive contractile organelles Algae as experimental systems Alan R. Liss Inc.,198919–37.
- 52.
- Schatten G. The centrosome and its mode of inheritance: the reduction of the centrosome during gametogenesis and its restoration during fertilization. Dev Biol. 1994;165:299–335. [PubMed: 7958403]
- 53.
- Khalfan W, Ivanovska I, Rose MD. Functional interaction between the PKC1 pathway and CDC31 network of SPB duplication genes. Genetics. 2000;155:1543–59. [PMC free article: PMC1461188] [PubMed: 10924456]
- 54.
- Ivanovska I, Rose MD. Fine structure analysis of the yeast centrin, Cdc31p, identifies residues specific for cell morphology and spindle pole body duplication. Genetics. 2001;157:503–18. [PMC free article: PMC1461518] [PubMed: 11156974]
- 55.
- Sullivan DS, Biggins S, Rose MD. The yeast centrin, Cdc31p, and interacting protein kinase, Kic1p, are required for cell integrity. J Cell Biol. 1998;143:751–65. [PMC free article: PMC2148137] [PubMed: 9813095]
- 56.
- Salisbury JL, Baron AT, Sanders MA. The centrin-based cytoskeleton of Chlamydomonas reinhardtii: distribution in interphase and mitotic cells. J Cell Biol. 1988;107:635–41. [PMC free article: PMC2115233] [PubMed: 3047144]
- 57.
- Baron AT, Salisbury JL. The centrin-related pericentriolar lattice of metazoan centrosomes. Comparative Spermatology 20 Years After. 1991;75:285–9.
- 58.
- Baron AT, Greenwood TM, Bazinet CW. et al. Centrin is a component of the pericentriolar lattice. Biol Cell. 1992;76:383–8. [PubMed: 1305481]
- 59.
- Laoukili J, Perret E, Middendorp S. et al. Differential expression and cellular distribution of centrin isoforms during human ciliated cell differentiation in vitro. J Cell Sci. 2000;113(8):1355–64. [PubMed: 10725219]
- 60.
- Paoletti A, Moudjou M, Paintrand M. et al. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J Cell Sci. 1996;109:3089–102. [PubMed: 9004043]
- 61.
- Middendorp S, Kuntziger T, Abraham Y. et al. A role for centrin 3 in centrosome reproduction. J Cell Biol. 2000;148(3):405–15. [PMC free article: PMC2174797] [PubMed: 10662768]
- 62.
- Besharse JC, Horst CJ. The photoreceptor connecting ciliuma model for the transition toneIn: Bloodgood RA, ed.Ciliary and Flagellar Membranes New York: Plenum,1990389–417.
- 63.
- Spencer M, Detwiler PB, Bunt-Milam AH. Distribution of membrane proteins in mechanical dissociated retinal rods. Invest Ophthalmol Visual Sci. 1988;29:1012–20. [PubMed: 2843476]
- 64.
- Wolfrum U, Schmitt A. Evidence for myosin VIIa driven rhodopsin transport in the plasma membrane of the photoreceptor connecting cilium In: Hollyfield JG, Andersson RE, LaVail M, eds.Retinal Degeneration Diseases and Experimental Therapy New York: Plenum Press,19993–14.
- 65.
- Wolfrum U, Schmitt A. Rhodopsin transport in the membrane of the connecting cilium of mammalian photoreceptor cells. Cell Motil Cytoskeleton. 2000;46:95–107. [PubMed: 10891855]
- 66.
- Liu X, Udovichenko IP, Brown SD. et al. Myosin VIIa participates in opsin transport through the photoreceptor cilium. J Neurosci. 1999;19(15):6267–74. [PMC free article: PMC6782817] [PubMed: 10414956]
- 67.
- Marszalek JR, Liu X, Roberts EA. et al. Genetic evidence for selective transport of opsin and arrestin by kinesin-II in mammalian photoreceptors. Cell. 2000;102:175–87. [PubMed: 10943838]
- 68.
- Uzawa M, Grams J, Madden B. et al. Identification of a complex between centrin and heat shock proteins in CSF-arrested Xenopus oocytes and dissociation of the complex following oocyte activation. Dev Biol. 1995;171:51–59. [PubMed: 7556907]
- 69.
- Paschke T. Untersuchungen zur Familie der Ca2+bindenden Centrine: biochemische Charakterisierung und Identifikation von interagierenden Proteinen Analysis of the family of Ca2+binding centrins: biochemical characterization and identification of interacting proteins Dissertation 1997. University of Cologne, Germany.
- 70.
- Philp NJ, Chang W, Long K. Light-stimulated protein movement in rod photoreceptor cells of the rat retina. FEBS Lett. 1987;225:127–32. [PubMed: 2826235]
- 71.
- Whelan JP, McGinnis JF. Light-dependent subcellular movement of photoreceptor proteins. J Neurosci Res. 1988;20:263–70. [PubMed: 3172281]
- 72.
- Matthews HR, Fain GL. A light-dependent increase in free Ca2+ concentration in the salamander rod outer segment. J Physiol. 2001;532:305–21. [PMC free article: PMC2278555] [PubMed: 11306652]
- 73.
- Bigay J, Faurobert E, Franco M. et al. Roles of lipid modifications of transducin subunits in their GDP-dependent association and membrane binding. Biochemistry. 1994;33:14081–90. [PubMed: 7947818]
Figures
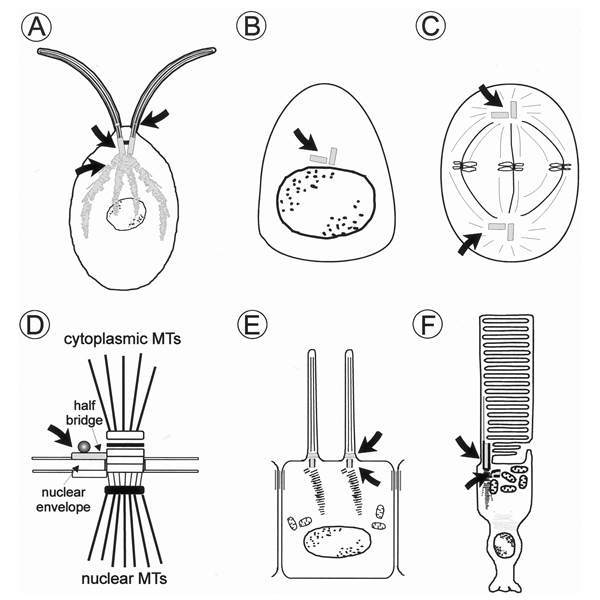
Figure 1
Localization of centrin in diverse cell types. Schematic diagrams of A unicellular green algae (e.g., Chlamydomonas reinhardtii); B animal cell in G1 or G0 phase (e.g., retinal nonphotoreceptor cells, cells of the retinal pigment epithelium) and C in metaphase; D spindle pole body of the yeast Saccharomyces cerevisiae, MTs = microtubules; E ciliated epidermal cell; F vertebrate photoreceptor cell. Centrin cellular localization is coloured and indicated by arrows. In the yeast, cdc31p (yeast centrin) is associated with the half bridge of the spindle pole body which accts as the major microtubule organizing centre (MTOC). Centrin is also commonly found at the MTOC, the centrosome of animal cells and at the centrosome-related basal bodies of ciliated cells. In cilia, centrin is also a component of the transition zone which links the basal body region with the axoneme.

Figure 2
Protein alignment of centrin isoforms and calmodulin from diverse species. ClustalX alignment of 15 different amino acid sequences of centrin species and rat calmodulin. (RnCaMp = rat calmodulin Accession Number (AN): CAA32120; NgCenp = Naegleria gruberi centrin AN: AAA75032; DsCenp = Dunaliella salina centrin AN: AAB67855; HsCen1p, 2p, 3p = human centrins 1, 2, 3 AN: AAC27343, AAH13873, AAH05383; MmCen1p, 3p = mouse centrins 1, 3 AN: AAD46390, AAH02162; RnCen1p, = rat centrins AN AAK20385, AnCenp = Atriplex nummularia centrin AN: P41210; CrCenp = Chlamydomonas reinhardtii centrin AN: CAA41039; SdCenp = Scherffelia dubia centrin AN: CAA49153; ScCdc31p = Saccharomyces cerevisiae (“yeast centrin”) AN: P06704; XlCenp = Xenopus laevis centrin AN: AAA79194; GiCenp = Giardia intestinalis centrin AN: AAB05594). EF-hand domains are indicated as a block above the sequence alignment. EF-hands are composed of an α-helix and a loop. Note, that the EF-hands 2 and 3 of most centrins appear most probably non-functional.
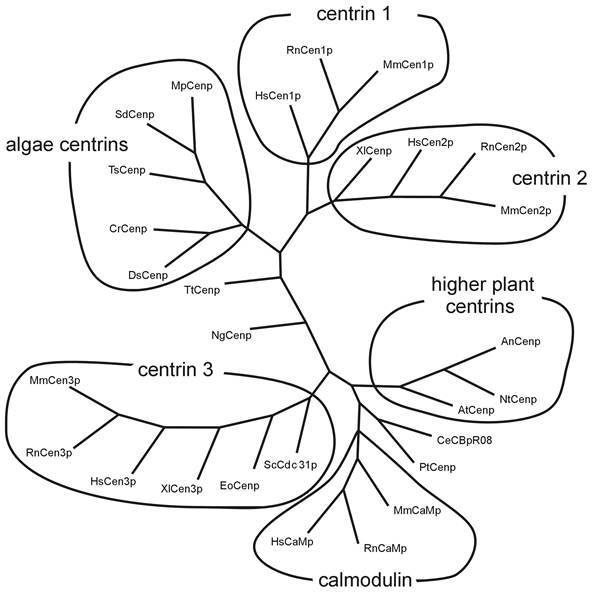
Figure 3
Comparison of centrin isoforms of diverse species. Comparison (using programs: Omiga 2.0, Genedoc 2.5.006 and phylip) of 28 different amino acid sequences of centrins and calmodulins. The phylogram shows a consensus tree which shows the highest frequency of each node of 1000 repetitions. Phylip divides the centrins into subgroups of centrin isoforms 1, 2, 3, algae centrins, higher plant centrins and a group of calmodulin (RnCaMp = rat calmodulin Accsession Number (AN): CAA32120; MmCaMp = mouse calmodulin AN: NP_033920; HsCaMp = human calmodulin AN: BAA08302; NgCenp = Naegleria gruberi centrin AN: AAA75032; XlCenp = Xenopus laevis centrin AN: AAA79194; XlCenp3 = Xenopus laevis centrin 3 AN AAG30507; PtCenp = Paramecium tetrauelia centrin AN: AAB188752; DsCenp = Dunaliella salina centrin AN: AAB67855; HsCen1p, 2p, 3p = human centrins 1, 2, 3 AN: AAC27343, AAH13873, AAH05383; MmCen1p, 2p, 3p = mouse centrins 1, 2, 3 AN: AAD46390, AAD46391, AAH02162; RnCen1p, 2p, 3p = rat centrins AN AAK20385, AAK20386, AAK83217; AtCenp = Arabidopsis thaliana centrin AN: CAB16762, AnCenp = Atriplex nummularia centrin AN: P41210; NtCenp = Nicotiana tabacum centrin AN AAF07221; CrCenp = Chlamydomonas reinhardtii centrin AN CAA41039; SdCenp = Scherffelia dubia centrin AN CAA49153; MpCenp = Micromonas pusilla centrin AN CAA58718; EoCenp = Euplotes octocarinatus centrin AN CAB40791; TsCenp = Tetraselmis striata centrin AN P43646; ScCdc31p = Saccharomyces cerevisiae AN P06704; CeCBpR08 = Caenorhabditis elegans AN P30644; TtCenp = Tetrahymena thermophila AN AAF66602. Tree is not complete.
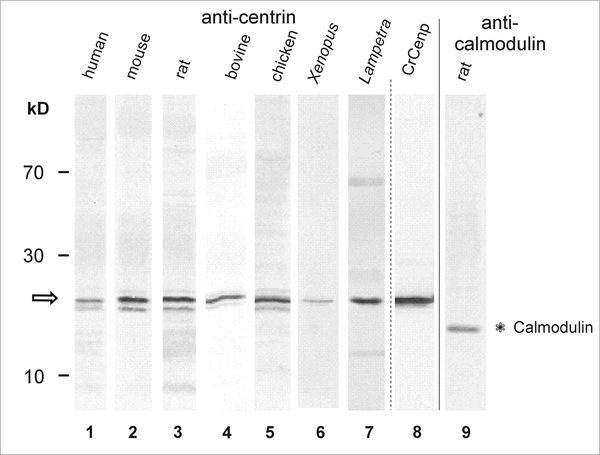
Figure 4
Western blot analysis reveals centrin expression in retina of various vertebrate species. Lane 1-Lane 8: Anti-centrin (mAb clone 20H5) Western blots. Lane 9: Anti-calmodulin Western blot of rat retina. Lane 1: human retina. Lane 2: mouse retina. Lane 3: rat retina. Lane 4: bovine retina. Lane 5: chicken retina. Lane 6: Xenopus retina. Lane 7: Lampetra retina. Lane 8: Bacterially expressed Chlamydomonas centrin. Anti-centrin antibodies detect bands at about the predicted molecular weight of 20 kDa (arrow) and do not crossreact with the calmodulin migrating at 17 kDa. Note: in some lanes (e.g., lane 5) several bands around 20 kDa are anti-centrin positive. These bands do neither represent different centrin isoforms nor different Ca2+-binding status of centrin. The higher bands most probably resemble phosphorylated centrin,43,44 and some lower bands may result from proteolytic digestion.
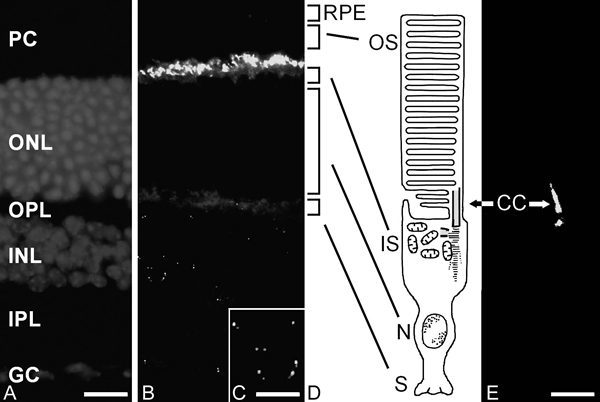
Figure 5
Localization of centrin in the mammalian retina and in photoreceptor fragments by indirect immunofluorescence. A DAPI-staining of a longitudinal cryosection through the rat retina. Staining of nuclei DNA demonstrates the retinal layers: PC = layer of outer and inner segments of photoreceptor cells; ONL = outer nuclear layer where nuclei of photoreceptors are localized; OPL = outer plexiform layer; INL = inner nuclear layer; IPL = inner plexiform layer; GC = ganglion cell layer. B Indirect anti-centrin immunofluorescence in the cryosection through rat retina. Anti-centrin antibodies predominantly react within the photoreceptor cell layer at the joint between the inner and outer segment of the photoreceptors. In addition, indirect immunofluorescence is present in dot pairs in the inner nuclear layer and ganglion cell layer. C Higher magnification of immunofluorescent staining with antibodies against centrin in the inner nuclear layer of the section shown in figure B. Centrin is present in the centrioles of the centrosomes present in the perikaryon of retinal neurons. Note that as a rule one centriole of a single centrosome shows brighter anti-centrin immunofluorescence. D Schematic representation of a mammalian rod photoreceptor cell. The light-sensitive outer segment (OS) is linked via the nonmotile connecting cilium (CC) with the inner segment (IS) where the protein synthesis machinery is localized. N = nuclear region; S = synaptic region. Centrin localization is indicated by the green color of centrin in the PRC. E Indirect anti-centrin immunofluorescence of a photoreceptor fragment of the rat retina analysed by confocal laser scanning microscopy. RPE = rod pigment epithelium. The figure shows the a labeling of the connecting cilium and the basal body.
Bars in B = A: 20 μm; C: 7 μm, E: 2 μm.
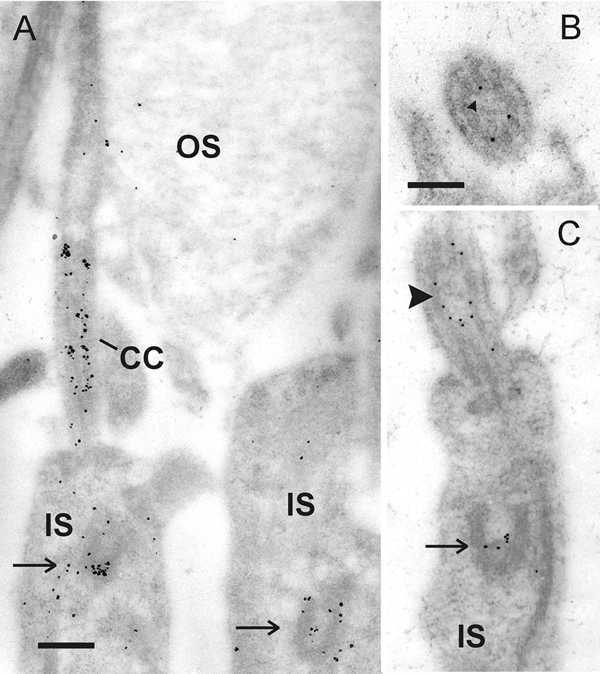
Figure 6
Immunoelectron microscopic localization of centrin in the connecting cilium of rod photoreceptor cells. A Silver-enhanced immunogold labeling of centrin in a longitudinal section of parts of rat rod photoreceptor cell. Centrin labeling is exclusively localized in the connecting cilium (CC) and the basal body complex (arrow) in the inner segment (IS) of photoreceptors. B Transversal section through the connecting cilium reveals that centrin is localized in the subciliary domain of the ciliary lumen encircled by axonemal microtubule doublets. C Slightly tangential section through the apical part of rat rod photoreceptor cell inner segment. Centrin antibodies react in the connecting cilium at the inner surface of the axonemal microtubule doublets (arrowhead). The arrow indicates basal body labeling.
Bars: A: 265 nm, B, C: 175 nm
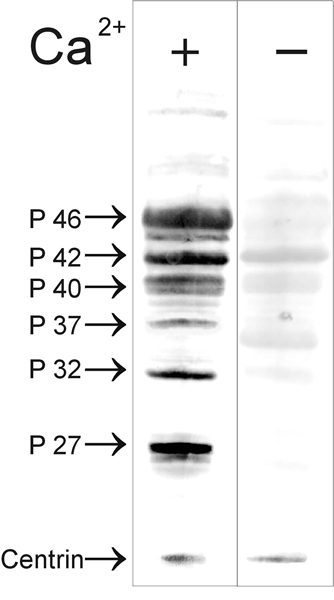
Figure 7
Analysis of centrin blot overlays with bovine retinal proteins. Western blot of retinal proteins overlay with recombinant expressed MmCen1p (67 μg/ml). Bound centrin was detected in a 2nd step by immunolabeling with anti-centrin antibodies (mAb clone 20H5). Lane 1: Centrin overlay assay in the presence of Ca2+ (1 mM CaCl2). Lane 2: Centrin overlay assay in the absence of Ca2+(6 mM EGtα). MmCen1p interaction with retinal proteins is dependent on the presence of Ca2+. In the absence of Ca2+, MmCen1p binding was dramatically reduced. Centrin interacting proteins are named according to their molecular weight (P 27, P 32, P 37, P 40, P 42, P46).
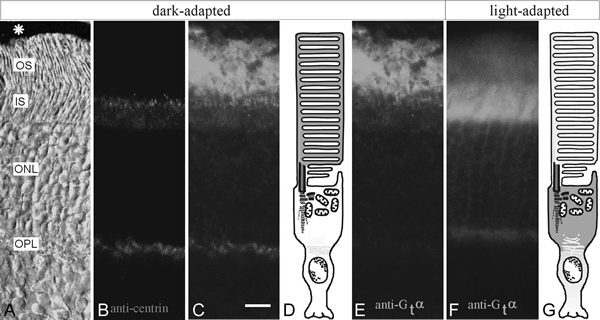
Figure 8
Light-dependent translocations of transducin in the mammalian retina. A-E dark-adapted mouse retina. F, G light-adapted retina. A DIC-image of a cryosection through mouse retina. Asterisk indicates retinal pigment epithelium, OS: photoreceptor outer segment; IS: photoreceptor inner segment, ONL: outer nuclear layer, OPL: outer plexiform layer. B anti-centrin immunofluorescence (Alexa,546) is concentrated in the connecting cilium between IS and OS of photoreceptor cells. C Merged images of B and E suggest partial co-localization of Gtα and centrin in the joint between both photoreceptor segments. D Schematic representation of a dark-adapted rod photoreceptor cell. Green colour indicates Gtα distribution. E Indirect anti-Gtα immunofluorescence in the double-labeled cryosection through the dark-adapted mouse retina shown in A-C. F Indirect anti-Gtα immunofluorescence in the section through the light-adapted mouse retina. G Schematic representation of a light-adapted rod photoreceptor cell. Green colour indicates Gtα distribution. In dark-adapted photoreceptor cells, Gtα is predominantly localized in the OS where as in the light-adapted condition Gtα is most prominent stained in the IS of photoreceptor cells.
Bar: 10 μm
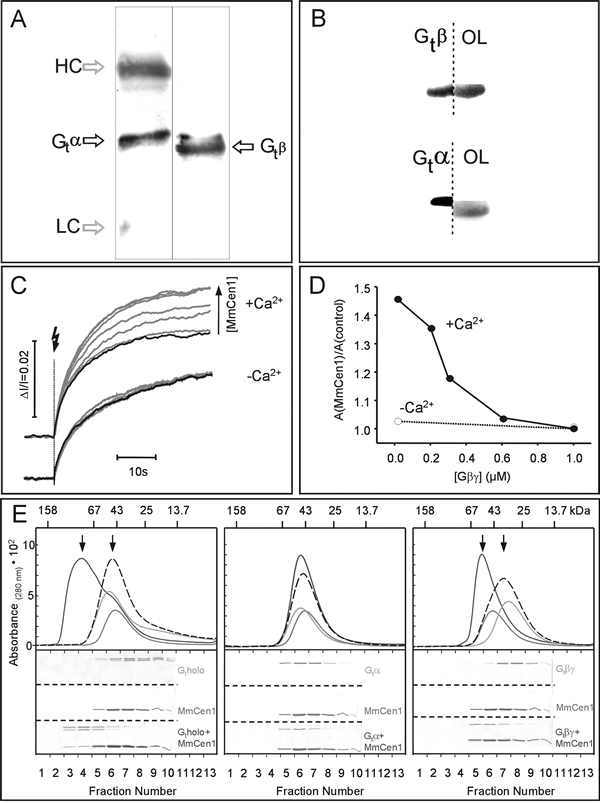
Figure 9
Calcium-dependent assembly of a centrin1/G-protein complex. A) Co-Immunoprecipitation of transducin with centrin from lysed retinal photoreceptor cell fragments.
Lanes 1: Western blot analysis with mAb anti-Gtα of an immunoprecipitation with mAb anti-centrin (clone 20H5) from photoreceptor cell fragments of bovine retina. (Upper and lower bands in lane 1 correspond to the heavy (HC) and light chains (LC) of mouse antibodies.) Lane 2: Western blot analysis with polyclonal anti-Gtβ of anti-centrin of an immunoprecipitation with mAb anti-centrin (clone 20H5) from photoreceptor cell fragments of bovine retina. Gtα and Gtβ co-immunoprecipitate with centrin. The upper and lower bands in lane 1 correspond to the heavy (HC) and light chains (LC) of the mouse antibodies.
B) Combined Western blot-overlay analysis identifies retinal centrin-interacting protein P37 as Gtβ subunit of transducin.
For specific determination of the centrin-binding protein, Western blotted lanes were cut in half and parallel processed for immunolabeling with subunit-specific antibodies against Gtβ (upper lane 1), and Gtα transducin (lower lane 1) and for overlays with recombinantly expressed MmCen1p (67 μg/ml) (OL). The 37 kDa centrin-binding protein (P37 in Fig. 7) is identified by centrin overlays had the exact mobility as the Gtβ subunit.
C) Calcium-dependent enhancement of kinetic light-scattering (KLS) Gt-binding signal in the presence of MmCen1p. Upper panel represents KLS binding signals (3 μM rhodopsin, 0.5 μM Gt) in the presence of calcium, and 0 (control, black curve), 0.6, 1.2, 2.5, 3.6, 5, 7.3, and 10 μM MmCen1p (gray curves), respectively. Lower panel represents KLS binding signals under the same conditions as in the upper panel, but with EGtα instead of calcium. Experimental conditions were 50 mM BTP, pH 7.5 containing 80 mM NaCl, 5 mM MgCl2 and either 100 μM CaCl2 or 1 mM EGtα at 20°C, sample volume of 300 μL, and cuvette path length of 1 cm; 32% rhodopsin was photolyzed per flash (500±20 nm).
D) Competition between Gtβγ-subunit and Gt for binding to MmCen1p. Calcium-dependent inhibition of the MmCen1p enhanced amplitude of flash-induced KLS Gt-binding signals by the βγ-subunit of Gt. The KLS assay was carried out as described in (A). Experiments were performed with the βγ-subunit of Gt. Data points represent the normalized amplitude of the MmCen1-dependent enhancement of the Gt-binding signal (AMmCen1) divided to the control Gt-binding signal without added MmCen1 (control). Filled and empty circles indicate the results obtained from experiments with calcium and with EGTA, respectively.
E) Calcium-dependent interaction of MmCen1p with Gt and its subunits analyzed by size-exclusion chromotography and SDS-PAGE.
Upper panels represent elution profiles of MmCen1p alone (...), Gt or its subunits alone (---), and the mixture of MmCen1p with Gt or its subunits (—) in the presence of calcium. The gray dotted lines are the calculated superpositions of the respective single component profiles (MmCen1p plus Gt or its subunits) yielding the predicted profiles for the mixture of the two noninteracting components. Arrows indicate the shift of the formed complexes. In the lower panel the SDS-PAGE analysis of the fractions of the size-exclusion chromatography is shown. Interaction of MmCen1p with the transducin holoprotein is shown in 1st panel with the Gtα-subunit in 2nd panel and with the Gtβγ-subunit in 3rd in the presence of calcium.
Experimental conditions: 10 μg of MmCen1p and 10 μg of Gtholo (or Gt subunits) were incubated in 50 mM BTP, pH 7.0 containing 80 mM NaCl, 1 mM MgCl2 and 100 μM CaCl2 for 5 min at room temperature, loaded on a Superose TM 12 column (using the Smart System, Pharmacia Biotech. Inc., flow rate, 40 μL/min) equilibrated with the same buffer, eluted by monitoring the absorbance at 280 nm and subsequent analyzed by SDS-PAGE. Note: Gt holoprotein elutes at an apparently lower MW, as compared to its subunits.73
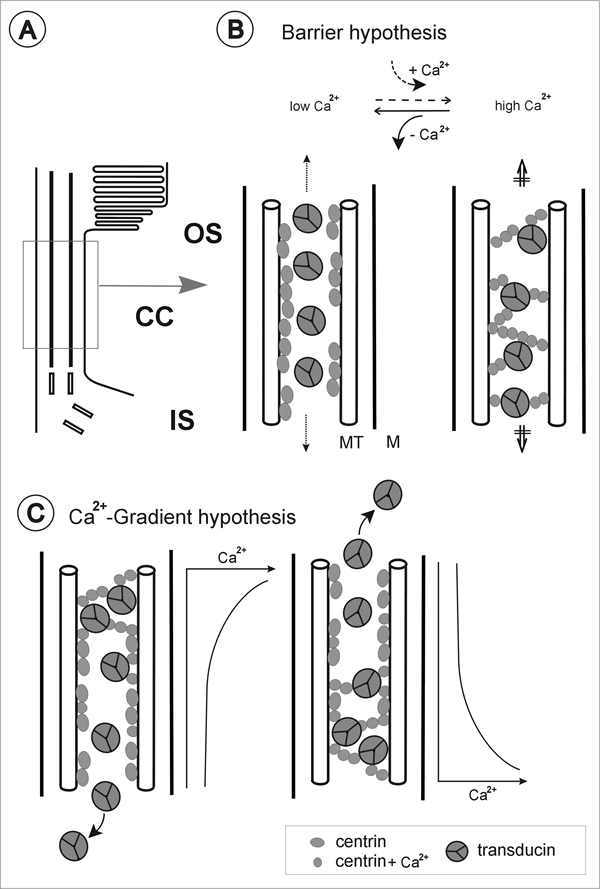
Figure 10
Models for Ca2+-dependent centrin-transducin assembly in the connecting cilium of vertebrate photoreceptor cell. A) Schematic representation of a part of a rod photoreceptor cell shows the linkage between the outer segment (OS) and inner segment (IS) by the connecting cilium (CC). (B and C) Enlargement of CC indicated in Figure A. B) Barrier hypothesis: under low free Ca2+ centrin is not activated and transducin floats through the inner lumen of the connecting cilium. If free Ca2+ increases in the outer segment and, in the cilium, centrin is activated by Ca2+ which induces Ca2+-centrin-transducin complex assembly and centrin fiber contraction. Thus transducin is trapped in the connecting cilium and a barrier between inner and outer segment raises. C) Ca2+-gradient hypothesis: transducin may bind to centrin 1 dependent on free Ca2+-concentration actually present in the ciliary domains. A putative Ca2+-gradient along the ciliary lumen may cause sequential assembly of the centrin 1-transducin complex and the release of transducin from the complex.